
Profundibacter amoris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Profundibacter
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
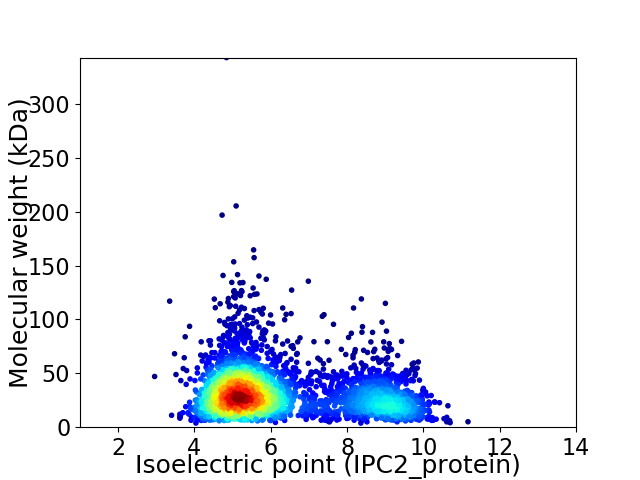
Virtual 2D-PAGE plot for 3454 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A347ULL6|A0A347ULL6_9RHOB DUF4198 domain-containing protein OS=Profundibacter amoris OX=2171755 GN=BAR1_08025 PE=4 SV=1
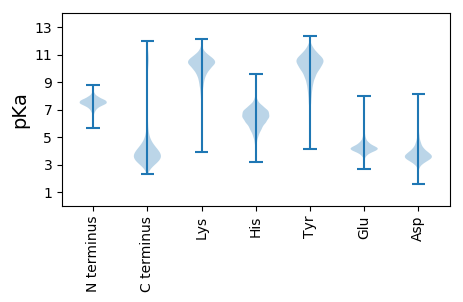
MM1 pKa = 7.83SDD3 pKa = 4.21NEE5 pKa = 4.66LNDD8 pKa = 3.95LDD10 pKa = 4.4GQISTFIADD19 pKa = 3.69EE20 pKa = 4.75LIAYY24 pKa = 9.04ADD26 pKa = 3.73QKK28 pKa = 11.49NVIIDD33 pKa = 3.78SIKK36 pKa = 10.38IGNEE40 pKa = 3.16YY41 pKa = 10.65DD42 pKa = 4.1LNGLSAAEE50 pKa = 4.05YY51 pKa = 9.57GQIASRR57 pKa = 11.84LSLIIGEE64 pKa = 4.69SLDD67 pKa = 3.67QYY69 pKa = 11.13GSSHH73 pKa = 6.57IGWGEE78 pKa = 3.58PKK80 pKa = 10.44LVVEE84 pKa = 5.38SGRR87 pKa = 11.84IWLQEE92 pKa = 4.01SGSGLFFGDD101 pKa = 3.44RR102 pKa = 11.84DD103 pKa = 3.99HH104 pKa = 7.11NGLMDD109 pKa = 3.89AQEE112 pKa = 4.31IKK114 pKa = 10.76DD115 pKa = 3.62AFDD118 pKa = 4.0TIEE121 pKa = 4.2AGYY124 pKa = 10.65VDD126 pKa = 4.75AVDD129 pKa = 3.38IHH131 pKa = 7.1SLTLFLSSYY140 pKa = 10.57EE141 pKa = 4.39DD142 pKa = 3.78YY143 pKa = 10.82FGQGPYY149 pKa = 11.03SNANLNTIYY158 pKa = 10.88SSLNSFWDD166 pKa = 3.44DD167 pKa = 3.11TFQDD171 pKa = 4.05VEE173 pKa = 4.38LHH175 pKa = 5.18TLAWQYY181 pKa = 10.18PWQRR185 pKa = 11.84DD186 pKa = 2.73IDD188 pKa = 4.2GPQGPGGVEE197 pKa = 3.85DD198 pKa = 4.28HH199 pKa = 6.83GGSALVNASLGFMQYY214 pKa = 11.1YY215 pKa = 9.85EE216 pKa = 4.16MSLGGVDD223 pKa = 4.73YY224 pKa = 10.84AVSWLASGWGGASPTLFHH242 pKa = 6.8NEE244 pKa = 3.28PRR246 pKa = 11.84AGGEE250 pKa = 4.06LFRR253 pKa = 11.84LMHH256 pKa = 6.25EE257 pKa = 4.7NISGLRR263 pKa = 11.84AIEE266 pKa = 4.16FDD268 pKa = 3.17QDD270 pKa = 3.27PTVFQNGDD278 pKa = 3.8DD279 pKa = 3.84IEE281 pKa = 5.07DD282 pKa = 3.49VVFRR286 pKa = 11.84AFEE289 pKa = 3.96QEE291 pKa = 4.23GKK293 pKa = 10.03VVLYY297 pKa = 10.27IGNLEE302 pKa = 4.35TEE304 pKa = 4.47SQTITIDD311 pKa = 3.95DD312 pKa = 3.74VLQFLEE318 pKa = 4.74GVDD321 pKa = 3.79GFGGFLNLNTTEE333 pKa = 4.99NIHH336 pKa = 5.62IWGSRR341 pKa = 11.84LGVDD345 pKa = 4.36GDD347 pKa = 4.15PNNYY351 pKa = 10.18SSMPVIDD358 pKa = 4.63IYY360 pKa = 11.69NHH362 pKa = 6.29IDD364 pKa = 3.41LFGDD368 pKa = 3.63SKK370 pKa = 11.68GFGALEE376 pKa = 4.17FEE378 pKa = 4.54LGAYY382 pKa = 9.89EE383 pKa = 4.79IMQLTFTSRR392 pKa = 11.84NHH394 pKa = 6.87DD395 pKa = 3.8LVEE398 pKa = 4.16MSGHH402 pKa = 6.28NGNDD406 pKa = 3.47TLIGSTWNDD415 pKa = 3.11EE416 pKa = 4.1LFGMGGNDD424 pKa = 3.77TIYY427 pKa = 11.08GGTGNDD433 pKa = 4.1AIYY436 pKa = 10.72GDD438 pKa = 4.25SGNDD442 pKa = 3.45EE443 pKa = 4.32LRR445 pKa = 11.84GDD447 pKa = 4.16AGNDD451 pKa = 3.58NISGGLGADD460 pKa = 3.69LVIGGTGNDD469 pKa = 3.88VISGGAYY476 pKa = 8.85GDD478 pKa = 3.73EE479 pKa = 4.31MFGGDD484 pKa = 4.54GFDD487 pKa = 5.09FINGGFGNDD496 pKa = 3.22RR497 pKa = 11.84LNGGEE502 pKa = 4.09GGDD505 pKa = 3.61KK506 pKa = 10.44FFHH509 pKa = 6.87LGIADD514 pKa = 4.56HH515 pKa = 6.66GSDD518 pKa = 4.31WIQDD522 pKa = 3.78YY523 pKa = 11.41NSVEE527 pKa = 4.07GDD529 pKa = 3.5VLLWGGGNATASDD542 pKa = 4.14FLVQRR547 pKa = 11.84AFTDD551 pKa = 3.46NAGDD555 pKa = 3.61ASVEE559 pKa = 4.37EE560 pKa = 4.44IFITHH565 pKa = 6.98IPTGNLLWALVDD577 pKa = 3.64GGEE580 pKa = 3.97QAEE583 pKa = 4.24INIQIGGEE591 pKa = 4.34VYY593 pKa = 10.94NLLL596 pKa = 4.23
MM1 pKa = 7.83SDD3 pKa = 4.21NEE5 pKa = 4.66LNDD8 pKa = 3.95LDD10 pKa = 4.4GQISTFIADD19 pKa = 3.69EE20 pKa = 4.75LIAYY24 pKa = 9.04ADD26 pKa = 3.73QKK28 pKa = 11.49NVIIDD33 pKa = 3.78SIKK36 pKa = 10.38IGNEE40 pKa = 3.16YY41 pKa = 10.65DD42 pKa = 4.1LNGLSAAEE50 pKa = 4.05YY51 pKa = 9.57GQIASRR57 pKa = 11.84LSLIIGEE64 pKa = 4.69SLDD67 pKa = 3.67QYY69 pKa = 11.13GSSHH73 pKa = 6.57IGWGEE78 pKa = 3.58PKK80 pKa = 10.44LVVEE84 pKa = 5.38SGRR87 pKa = 11.84IWLQEE92 pKa = 4.01SGSGLFFGDD101 pKa = 3.44RR102 pKa = 11.84DD103 pKa = 3.99HH104 pKa = 7.11NGLMDD109 pKa = 3.89AQEE112 pKa = 4.31IKK114 pKa = 10.76DD115 pKa = 3.62AFDD118 pKa = 4.0TIEE121 pKa = 4.2AGYY124 pKa = 10.65VDD126 pKa = 4.75AVDD129 pKa = 3.38IHH131 pKa = 7.1SLTLFLSSYY140 pKa = 10.57EE141 pKa = 4.39DD142 pKa = 3.78YY143 pKa = 10.82FGQGPYY149 pKa = 11.03SNANLNTIYY158 pKa = 10.88SSLNSFWDD166 pKa = 3.44DD167 pKa = 3.11TFQDD171 pKa = 4.05VEE173 pKa = 4.38LHH175 pKa = 5.18TLAWQYY181 pKa = 10.18PWQRR185 pKa = 11.84DD186 pKa = 2.73IDD188 pKa = 4.2GPQGPGGVEE197 pKa = 3.85DD198 pKa = 4.28HH199 pKa = 6.83GGSALVNASLGFMQYY214 pKa = 11.1YY215 pKa = 9.85EE216 pKa = 4.16MSLGGVDD223 pKa = 4.73YY224 pKa = 10.84AVSWLASGWGGASPTLFHH242 pKa = 6.8NEE244 pKa = 3.28PRR246 pKa = 11.84AGGEE250 pKa = 4.06LFRR253 pKa = 11.84LMHH256 pKa = 6.25EE257 pKa = 4.7NISGLRR263 pKa = 11.84AIEE266 pKa = 4.16FDD268 pKa = 3.17QDD270 pKa = 3.27PTVFQNGDD278 pKa = 3.8DD279 pKa = 3.84IEE281 pKa = 5.07DD282 pKa = 3.49VVFRR286 pKa = 11.84AFEE289 pKa = 3.96QEE291 pKa = 4.23GKK293 pKa = 10.03VVLYY297 pKa = 10.27IGNLEE302 pKa = 4.35TEE304 pKa = 4.47SQTITIDD311 pKa = 3.95DD312 pKa = 3.74VLQFLEE318 pKa = 4.74GVDD321 pKa = 3.79GFGGFLNLNTTEE333 pKa = 4.99NIHH336 pKa = 5.62IWGSRR341 pKa = 11.84LGVDD345 pKa = 4.36GDD347 pKa = 4.15PNNYY351 pKa = 10.18SSMPVIDD358 pKa = 4.63IYY360 pKa = 11.69NHH362 pKa = 6.29IDD364 pKa = 3.41LFGDD368 pKa = 3.63SKK370 pKa = 11.68GFGALEE376 pKa = 4.17FEE378 pKa = 4.54LGAYY382 pKa = 9.89EE383 pKa = 4.79IMQLTFTSRR392 pKa = 11.84NHH394 pKa = 6.87DD395 pKa = 3.8LVEE398 pKa = 4.16MSGHH402 pKa = 6.28NGNDD406 pKa = 3.47TLIGSTWNDD415 pKa = 3.11EE416 pKa = 4.1LFGMGGNDD424 pKa = 3.77TIYY427 pKa = 11.08GGTGNDD433 pKa = 4.1AIYY436 pKa = 10.72GDD438 pKa = 4.25SGNDD442 pKa = 3.45EE443 pKa = 4.32LRR445 pKa = 11.84GDD447 pKa = 4.16AGNDD451 pKa = 3.58NISGGLGADD460 pKa = 3.69LVIGGTGNDD469 pKa = 3.88VISGGAYY476 pKa = 8.85GDD478 pKa = 3.73EE479 pKa = 4.31MFGGDD484 pKa = 4.54GFDD487 pKa = 5.09FINGGFGNDD496 pKa = 3.22RR497 pKa = 11.84LNGGEE502 pKa = 4.09GGDD505 pKa = 3.61KK506 pKa = 10.44FFHH509 pKa = 6.87LGIADD514 pKa = 4.56HH515 pKa = 6.66GSDD518 pKa = 4.31WIQDD522 pKa = 3.78YY523 pKa = 11.41NSVEE527 pKa = 4.07GDD529 pKa = 3.5VLLWGGGNATASDD542 pKa = 4.14FLVQRR547 pKa = 11.84AFTDD551 pKa = 3.46NAGDD555 pKa = 3.61ASVEE559 pKa = 4.37EE560 pKa = 4.44IFITHH565 pKa = 6.98IPTGNLLWALVDD577 pKa = 3.64GGEE580 pKa = 3.97QAEE583 pKa = 4.24INIQIGGEE591 pKa = 4.34VYY593 pKa = 10.94NLLL596 pKa = 4.23
Molecular weight: 64.46 kDa
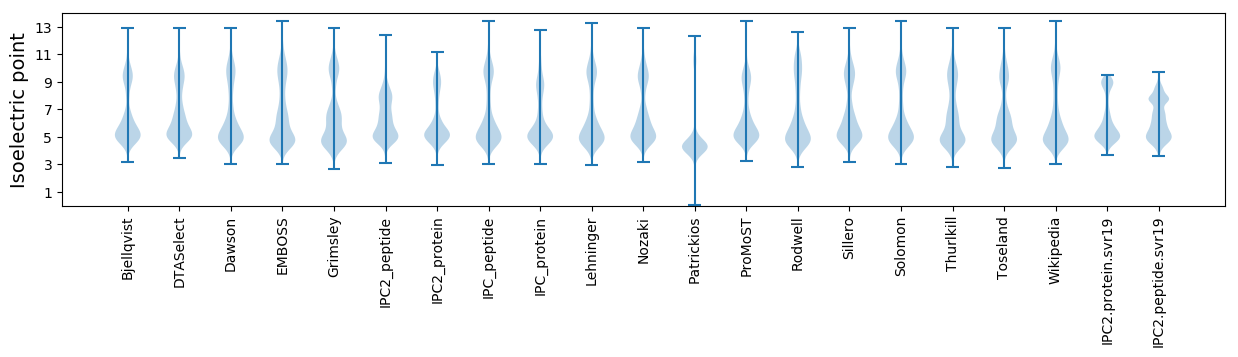
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A347UCR7|A0A347UCR7_9RHOB Thiamine/thiamine pyrophosphate ABC transporter permease ThiP OS=Profundibacter amoris OX=2171755 GN=BAR1_01035 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.83HH14 pKa = 4.64RR15 pKa = 11.84HH16 pKa = 3.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.83HH14 pKa = 4.64RR15 pKa = 11.84HH16 pKa = 3.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1063791 |
35 |
3278 |
308.0 |
33.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.254 ± 0.045 | 0.889 ± 0.014 |
6.114 ± 0.034 | 5.835 ± 0.036 |
3.868 ± 0.031 | 8.468 ± 0.044 |
2.005 ± 0.022 | 5.697 ± 0.03 |
4.169 ± 0.036 | 9.815 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.987 ± 0.024 | 3.06 ± 0.025 |
4.869 ± 0.03 | 3.279 ± 0.023 |
6.056 ± 0.037 | 5.069 ± 0.029 |
5.515 ± 0.031 | 7.22 ± 0.038 |
1.373 ± 0.019 | 2.46 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
