
Apis mellifera associated microvirus 41
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 8.0
Get precalculated fractions of proteins
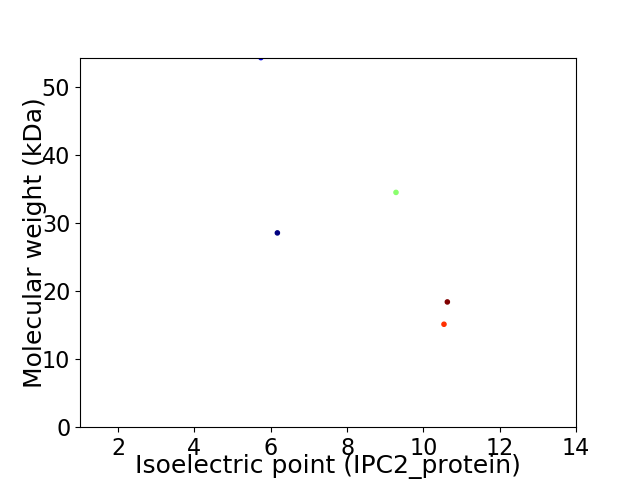
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
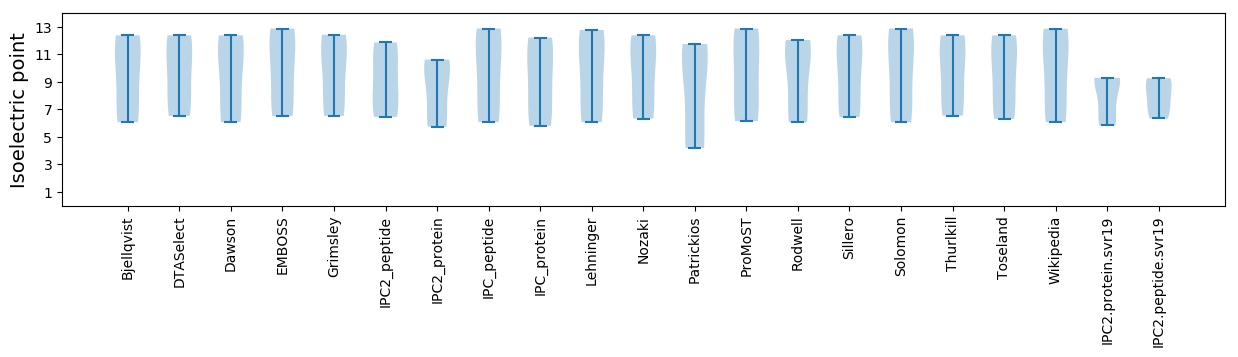
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3Q8U5W2|A0A3Q8U5W2_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 41 OX=2494771 PE=4 SV=1
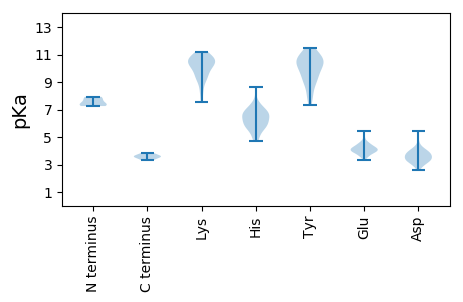
MM1 pKa = 7.25KK2 pKa = 10.06RR3 pKa = 11.84SKK5 pKa = 10.7FSLSHH10 pKa = 6.26YY11 pKa = 10.42KK12 pKa = 10.54LLSCDD17 pKa = 3.28MGEE20 pKa = 5.45LIPCGLQEE28 pKa = 4.61VLPGDD33 pKa = 4.24TFQQSTSILVRR44 pKa = 11.84ASPLLAPVMHH54 pKa = 6.83PVHH57 pKa = 6.76VRR59 pKa = 11.84IHH61 pKa = 5.67HH62 pKa = 5.87WFVPHH67 pKa = 6.64RR68 pKa = 11.84LVWEE72 pKa = 4.18DD73 pKa = 2.96WEE75 pKa = 4.29EE76 pKa = 4.51FITGGPDD83 pKa = 3.24GMDD86 pKa = 3.17DD87 pKa = 4.07SVFPTITAGGSGYY100 pKa = 9.79PVGSLSDD107 pKa = 3.83YY108 pKa = 10.94LGVPPGVANLEE119 pKa = 4.43HH120 pKa = 6.74SALPHH125 pKa = 6.38RR126 pKa = 11.84GYY128 pKa = 11.51GLIWNEE134 pKa = 3.74WYY136 pKa = 10.21RR137 pKa = 11.84DD138 pKa = 3.16QDD140 pKa = 3.6LQSEE144 pKa = 4.79VFVDD148 pKa = 3.75KK149 pKa = 11.06TSGPDD154 pKa = 3.2ANTSVALQNVGWEE167 pKa = 3.99KK168 pKa = 11.19DD169 pKa = 3.66YY170 pKa = 9.37FTSARR175 pKa = 11.84PWEE178 pKa = 4.41QKK180 pKa = 10.76GPTISIPVGGAAPVTGSVEE199 pKa = 4.04FLTNSTVRR207 pKa = 11.84TVSATGGTSGARR219 pKa = 11.84GMEE222 pKa = 3.77IPATNNPDD230 pKa = 3.11TVMARR235 pKa = 11.84LGADD239 pKa = 3.28AEE241 pKa = 4.56ADD243 pKa = 3.52LSGASAVNVNILRR256 pKa = 11.84EE257 pKa = 4.0ALALQRR263 pKa = 11.84YY264 pKa = 8.18EE265 pKa = 4.12EE266 pKa = 3.92ARR268 pKa = 11.84ARR270 pKa = 11.84YY271 pKa = 7.87GSRR274 pKa = 11.84YY275 pKa = 9.33VEE277 pKa = 3.92YY278 pKa = 10.67LRR280 pKa = 11.84YY281 pKa = 10.05LGVRR285 pKa = 11.84SSDD288 pKa = 3.15ARR290 pKa = 11.84LQRR293 pKa = 11.84PEE295 pKa = 3.94YY296 pKa = 10.49LGGGQATIQFSEE308 pKa = 4.39VLQTAEE314 pKa = 3.79GTNPVGEE321 pKa = 4.25LRR323 pKa = 11.84GHH325 pKa = 7.28GIGTPRR331 pKa = 11.84SNRR334 pKa = 11.84YY335 pKa = 8.58RR336 pKa = 11.84KK337 pKa = 9.26FFEE340 pKa = 3.63EE341 pKa = 3.66HH342 pKa = 6.59GYY344 pKa = 8.97IHH346 pKa = 6.6TFVSVRR352 pKa = 11.84PKK354 pKa = 9.96TIYY357 pKa = 10.75ANGLFRR363 pKa = 11.84HH364 pKa = 5.65WNRR367 pKa = 11.84RR368 pKa = 11.84TKK370 pKa = 10.55EE371 pKa = 4.11DD372 pKa = 2.95FWQRR376 pKa = 11.84EE377 pKa = 4.21LQHH380 pKa = 6.98IGQQEE385 pKa = 4.19VLNKK389 pKa = 9.71EE390 pKa = 4.23VYY392 pKa = 9.69AASASPDD399 pKa = 3.61DD400 pKa = 3.73VFGFQDD406 pKa = 3.41RR407 pKa = 11.84YY408 pKa = 10.77DD409 pKa = 3.49EE410 pKa = 4.06YY411 pKa = 11.43RR412 pKa = 11.84RR413 pKa = 11.84TEE415 pKa = 3.84SSIGGEE421 pKa = 4.02FRR423 pKa = 11.84TTLDD427 pKa = 3.33FWHH430 pKa = 7.02MARR433 pKa = 11.84IFSSSPALNADD444 pKa = 3.81FVKK447 pKa = 10.63SVPTEE452 pKa = 3.61RR453 pKa = 11.84TFAVPSEE460 pKa = 4.06DD461 pKa = 4.12VLWLMARR468 pKa = 11.84HH469 pKa = 6.41NIQARR474 pKa = 11.84RR475 pKa = 11.84MVAAQGHH482 pKa = 5.72SYY484 pKa = 10.71IFF486 pKa = 3.83
MM1 pKa = 7.25KK2 pKa = 10.06RR3 pKa = 11.84SKK5 pKa = 10.7FSLSHH10 pKa = 6.26YY11 pKa = 10.42KK12 pKa = 10.54LLSCDD17 pKa = 3.28MGEE20 pKa = 5.45LIPCGLQEE28 pKa = 4.61VLPGDD33 pKa = 4.24TFQQSTSILVRR44 pKa = 11.84ASPLLAPVMHH54 pKa = 6.83PVHH57 pKa = 6.76VRR59 pKa = 11.84IHH61 pKa = 5.67HH62 pKa = 5.87WFVPHH67 pKa = 6.64RR68 pKa = 11.84LVWEE72 pKa = 4.18DD73 pKa = 2.96WEE75 pKa = 4.29EE76 pKa = 4.51FITGGPDD83 pKa = 3.24GMDD86 pKa = 3.17DD87 pKa = 4.07SVFPTITAGGSGYY100 pKa = 9.79PVGSLSDD107 pKa = 3.83YY108 pKa = 10.94LGVPPGVANLEE119 pKa = 4.43HH120 pKa = 6.74SALPHH125 pKa = 6.38RR126 pKa = 11.84GYY128 pKa = 11.51GLIWNEE134 pKa = 3.74WYY136 pKa = 10.21RR137 pKa = 11.84DD138 pKa = 3.16QDD140 pKa = 3.6LQSEE144 pKa = 4.79VFVDD148 pKa = 3.75KK149 pKa = 11.06TSGPDD154 pKa = 3.2ANTSVALQNVGWEE167 pKa = 3.99KK168 pKa = 11.19DD169 pKa = 3.66YY170 pKa = 9.37FTSARR175 pKa = 11.84PWEE178 pKa = 4.41QKK180 pKa = 10.76GPTISIPVGGAAPVTGSVEE199 pKa = 4.04FLTNSTVRR207 pKa = 11.84TVSATGGTSGARR219 pKa = 11.84GMEE222 pKa = 3.77IPATNNPDD230 pKa = 3.11TVMARR235 pKa = 11.84LGADD239 pKa = 3.28AEE241 pKa = 4.56ADD243 pKa = 3.52LSGASAVNVNILRR256 pKa = 11.84EE257 pKa = 4.0ALALQRR263 pKa = 11.84YY264 pKa = 8.18EE265 pKa = 4.12EE266 pKa = 3.92ARR268 pKa = 11.84ARR270 pKa = 11.84YY271 pKa = 7.87GSRR274 pKa = 11.84YY275 pKa = 9.33VEE277 pKa = 3.92YY278 pKa = 10.67LRR280 pKa = 11.84YY281 pKa = 10.05LGVRR285 pKa = 11.84SSDD288 pKa = 3.15ARR290 pKa = 11.84LQRR293 pKa = 11.84PEE295 pKa = 3.94YY296 pKa = 10.49LGGGQATIQFSEE308 pKa = 4.39VLQTAEE314 pKa = 3.79GTNPVGEE321 pKa = 4.25LRR323 pKa = 11.84GHH325 pKa = 7.28GIGTPRR331 pKa = 11.84SNRR334 pKa = 11.84YY335 pKa = 8.58RR336 pKa = 11.84KK337 pKa = 9.26FFEE340 pKa = 3.63EE341 pKa = 3.66HH342 pKa = 6.59GYY344 pKa = 8.97IHH346 pKa = 6.6TFVSVRR352 pKa = 11.84PKK354 pKa = 9.96TIYY357 pKa = 10.75ANGLFRR363 pKa = 11.84HH364 pKa = 5.65WNRR367 pKa = 11.84RR368 pKa = 11.84TKK370 pKa = 10.55EE371 pKa = 4.11DD372 pKa = 2.95FWQRR376 pKa = 11.84EE377 pKa = 4.21LQHH380 pKa = 6.98IGQQEE385 pKa = 4.19VLNKK389 pKa = 9.71EE390 pKa = 4.23VYY392 pKa = 9.69AASASPDD399 pKa = 3.61DD400 pKa = 3.73VFGFQDD406 pKa = 3.41RR407 pKa = 11.84YY408 pKa = 10.77DD409 pKa = 3.49EE410 pKa = 4.06YY411 pKa = 11.43RR412 pKa = 11.84RR413 pKa = 11.84TEE415 pKa = 3.84SSIGGEE421 pKa = 4.02FRR423 pKa = 11.84TTLDD427 pKa = 3.33FWHH430 pKa = 7.02MARR433 pKa = 11.84IFSSSPALNADD444 pKa = 3.81FVKK447 pKa = 10.63SVPTEE452 pKa = 3.61RR453 pKa = 11.84TFAVPSEE460 pKa = 4.06DD461 pKa = 4.12VLWLMARR468 pKa = 11.84HH469 pKa = 6.41NIQARR474 pKa = 11.84RR475 pKa = 11.84MVAAQGHH482 pKa = 5.72SYY484 pKa = 10.71IFF486 pKa = 3.83
Molecular weight: 54.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U5L1|A0A3Q8U5L1_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 41 OX=2494771 PE=3 SV=1
MM1 pKa = 7.93AARR4 pKa = 11.84QAQPRR9 pKa = 11.84GCKK12 pKa = 9.27LCKK15 pKa = 10.12KK16 pKa = 8.53ITPSTTWMASRR27 pKa = 11.84FMMSGDD33 pKa = 3.44MKK35 pKa = 11.11SPIPRR40 pKa = 11.84RR41 pKa = 11.84SRR43 pKa = 11.84QRR45 pKa = 11.84SATSVSRR52 pKa = 11.84LSLTKK57 pKa = 10.13SGQWSGLNVSPRR69 pKa = 11.84KK70 pKa = 7.98RR71 pKa = 11.84QRR73 pKa = 11.84WALRR77 pKa = 11.84PSKK80 pKa = 10.43RR81 pKa = 11.84PTISRR86 pKa = 11.84SAIRR90 pKa = 11.84MTSFQALHH98 pKa = 6.57TSTISTPCPCPSSRR112 pKa = 11.84LVRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84PKK119 pKa = 10.5RR120 pKa = 11.84PTRR123 pKa = 11.84LPLQSPNLRR132 pKa = 11.84KK133 pKa = 9.84NLATTTSRR141 pKa = 11.84RR142 pKa = 11.84LHH144 pKa = 5.87LRR146 pKa = 11.84RR147 pKa = 11.84EE148 pKa = 3.92RR149 pKa = 11.84DD150 pKa = 3.43LANSPAPVGAAPSAA164 pKa = 3.57
MM1 pKa = 7.93AARR4 pKa = 11.84QAQPRR9 pKa = 11.84GCKK12 pKa = 9.27LCKK15 pKa = 10.12KK16 pKa = 8.53ITPSTTWMASRR27 pKa = 11.84FMMSGDD33 pKa = 3.44MKK35 pKa = 11.11SPIPRR40 pKa = 11.84RR41 pKa = 11.84SRR43 pKa = 11.84QRR45 pKa = 11.84SATSVSRR52 pKa = 11.84LSLTKK57 pKa = 10.13SGQWSGLNVSPRR69 pKa = 11.84KK70 pKa = 7.98RR71 pKa = 11.84QRR73 pKa = 11.84WALRR77 pKa = 11.84PSKK80 pKa = 10.43RR81 pKa = 11.84PTISRR86 pKa = 11.84SAIRR90 pKa = 11.84MTSFQALHH98 pKa = 6.57TSTISTPCPCPSSRR112 pKa = 11.84LVRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84PKK119 pKa = 10.5RR120 pKa = 11.84PTRR123 pKa = 11.84LPLQSPNLRR132 pKa = 11.84KK133 pKa = 9.84NLATTTSRR141 pKa = 11.84RR142 pKa = 11.84LHH144 pKa = 5.87LRR146 pKa = 11.84RR147 pKa = 11.84EE148 pKa = 3.92RR149 pKa = 11.84DD150 pKa = 3.43LANSPAPVGAAPSAA164 pKa = 3.57
Molecular weight: 18.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1352 |
133 |
486 |
270.4 |
30.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.58 ± 0.805 | 1.331 ± 0.673 |
4.29 ± 0.789 | 4.438 ± 1.01 |
3.402 ± 0.538 | 7.618 ± 1.174 |
2.589 ± 0.35 | 3.254 ± 0.267 |
3.624 ± 0.782 | 8.136 ± 0.769 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.515 ± 0.332 | 3.033 ± 0.247 |
6.065 ± 0.805 | 4.438 ± 0.912 |
10.429 ± 1.918 | 9.467 ± 1.476 |
5.399 ± 0.786 | 6.731 ± 0.942 |
1.701 ± 0.23 | 2.959 ± 0.597 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
