
Pseudomonas indica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
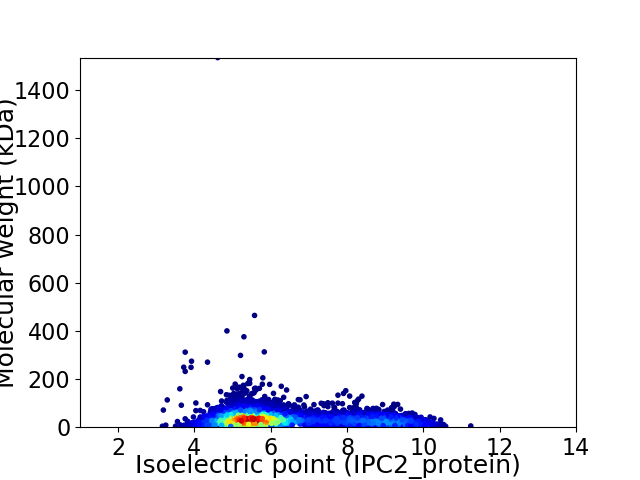
Virtual 2D-PAGE plot for 5180 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8U1M2|A0A1G8U1M2_9PSED Phosphate-specific transport system accessory protein PhoU OS=Pseudomonas indica OX=137658 GN=SAMN05216186_101522 PE=3 SV=1
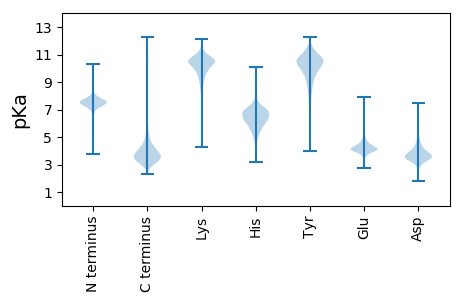
MM1 pKa = 7.32HH2 pKa = 7.16THH4 pKa = 6.3QLLQSLPRR12 pKa = 11.84CLLSAALVAGMASTAGVYY30 pKa = 10.17AVDD33 pKa = 4.37GAPPGLFEE41 pKa = 6.68LDD43 pKa = 3.42GNTVDD48 pKa = 5.0GAAAGDD54 pKa = 3.82DD55 pKa = 3.71WGSLHH60 pKa = 7.12LGNQAGSPVAFTGILSDD77 pKa = 3.88PAPVTIYY84 pKa = 9.67TQGGSKK90 pKa = 10.34DD91 pKa = 3.94VNDD94 pKa = 3.58VTQWRR99 pKa = 11.84YY100 pKa = 8.04TNGSVPDD107 pKa = 4.25KK108 pKa = 11.35DD109 pKa = 5.71DD110 pKa = 3.1ITNAYY115 pKa = 7.15ATAYY119 pKa = 9.52INPADD124 pKa = 3.82VSHH127 pKa = 6.82NGQVVHH133 pKa = 6.84AAGDD137 pKa = 3.96LIIYY141 pKa = 9.66FGLDD145 pKa = 2.55RR146 pKa = 11.84FANNGDD152 pKa = 3.32AFAGFWFFQDD162 pKa = 3.73NVTLGANGRR171 pKa = 11.84FNGQHH176 pKa = 5.6VARR179 pKa = 11.84QGMEE183 pKa = 3.9GQPGYY188 pKa = 11.31VPGDD192 pKa = 3.42LLVLVEE198 pKa = 4.33YY199 pKa = 9.55PQGANAQPEE208 pKa = 4.43IKK210 pKa = 10.43VYY212 pKa = 10.22EE213 pKa = 4.32WDD215 pKa = 4.47PLDD218 pKa = 5.09LDD220 pKa = 4.53HH221 pKa = 8.09DD222 pKa = 4.33NVAPNLDD229 pKa = 3.39QRR231 pKa = 11.84YY232 pKa = 9.18SSAAAKK238 pKa = 10.44CDD240 pKa = 3.33GSGNKK245 pKa = 8.66LACAITNVANLPGAPEE261 pKa = 3.77WPYY264 pKa = 10.01TPKK267 pKa = 10.8NGFAGIPFEE276 pKa = 4.59SFFEE280 pKa = 4.27GGINVSKK287 pKa = 10.9LLGGDD292 pKa = 3.68VPCFSSFLAEE302 pKa = 4.17TRR304 pKa = 11.84SSRR307 pKa = 11.84SEE309 pKa = 3.78TSQLKK314 pKa = 10.83DD315 pKa = 3.52FVLGDD320 pKa = 3.95FNLCSIAVTKK330 pKa = 10.62QCTAAVDD337 pKa = 3.65AGDD340 pKa = 4.24GGNSVLVNYY349 pKa = 10.51SGTVTNDD356 pKa = 2.7GGLPLFDD363 pKa = 3.5VTVTDD368 pKa = 5.9DD369 pKa = 3.2MGTANTADD377 pKa = 3.63DD378 pKa = 3.95VVVFGPATLQPGEE391 pKa = 4.22SQPYY395 pKa = 7.63NGSYY399 pKa = 8.85STLSIPATDD408 pKa = 3.36EE409 pKa = 3.91VTAVGHH415 pKa = 6.44RR416 pKa = 11.84NGTSVEE422 pKa = 4.05ATAEE426 pKa = 4.34ASCSPDD432 pKa = 3.21VTPALAVDD440 pKa = 4.54KK441 pKa = 11.2ACTANIASGGAGIDD455 pKa = 3.29VLFNGTVTNTGNVALEE471 pKa = 4.0NVMVVDD477 pKa = 5.49DD478 pKa = 5.18GGTPGDD484 pKa = 3.73TSDD487 pKa = 4.36DD488 pKa = 3.86VTVLGPVTLNAGEE501 pKa = 4.17SMPYY505 pKa = 9.87NGSFSASGSNSSTDD519 pKa = 3.15TVTATGSDD527 pKa = 3.85VLTQSPVEE535 pKa = 4.03ATAQATCEE543 pKa = 4.05ADD545 pKa = 3.3VTPAIEE551 pKa = 4.2VTKK554 pKa = 10.67QCSANINTGGTGIDD568 pKa = 3.32VLFTGTVTNTGDD580 pKa = 3.25VALGNVTVVDD590 pKa = 4.43DD591 pKa = 4.55NGTPGDD597 pKa = 3.76TSDD600 pKa = 4.36DD601 pKa = 3.85VTVLGPISLAPGEE614 pKa = 4.26SANYY618 pKa = 9.5NGSFSASGSSSTDD631 pKa = 2.84TVTASGADD639 pKa = 3.59VLTQSPVEE647 pKa = 4.12AQASASCSADD657 pKa = 3.04VTPAIEE663 pKa = 4.21VTKK666 pKa = 10.74QCTDD670 pKa = 2.93APAFGQAILFDD681 pKa = 3.82GTVTNTGNVALLNVTVVDD699 pKa = 4.68DD700 pKa = 4.4NGTPGDD706 pKa = 3.81TSDD709 pKa = 5.61DD710 pKa = 3.62VTFNLGDD717 pKa = 4.13LAPGAAANYY726 pKa = 9.03NGAYY730 pKa = 8.14TPSLAGSYY738 pKa = 9.71TNTVVATAEE747 pKa = 4.19DD748 pKa = 4.14AVEE751 pKa = 4.07SSPVSSSADD760 pKa = 3.35ATCEE764 pKa = 3.99VPPPPEE770 pKa = 5.48GEE772 pKa = 4.16GCTPGFWKK780 pKa = 10.55NSPGSWGPTGYY791 pKa = 10.55SPNQTVGSVFSLPSGQLANQLGGNSLIQALNYY823 pKa = 10.69AGGNNLLGSAQILLRR838 pKa = 11.84AAVAAVLNASHH849 pKa = 7.71PDD851 pKa = 2.81VDD853 pKa = 4.28YY854 pKa = 10.37PRR856 pKa = 11.84TAASVIADD864 pKa = 3.58VNAALATKK872 pKa = 10.43DD873 pKa = 3.17RR874 pKa = 11.84ATILALAAALDD885 pKa = 3.9ADD887 pKa = 4.3NNLGCDD893 pKa = 3.99LPNDD897 pKa = 3.53NSFF900 pKa = 3.53
MM1 pKa = 7.32HH2 pKa = 7.16THH4 pKa = 6.3QLLQSLPRR12 pKa = 11.84CLLSAALVAGMASTAGVYY30 pKa = 10.17AVDD33 pKa = 4.37GAPPGLFEE41 pKa = 6.68LDD43 pKa = 3.42GNTVDD48 pKa = 5.0GAAAGDD54 pKa = 3.82DD55 pKa = 3.71WGSLHH60 pKa = 7.12LGNQAGSPVAFTGILSDD77 pKa = 3.88PAPVTIYY84 pKa = 9.67TQGGSKK90 pKa = 10.34DD91 pKa = 3.94VNDD94 pKa = 3.58VTQWRR99 pKa = 11.84YY100 pKa = 8.04TNGSVPDD107 pKa = 4.25KK108 pKa = 11.35DD109 pKa = 5.71DD110 pKa = 3.1ITNAYY115 pKa = 7.15ATAYY119 pKa = 9.52INPADD124 pKa = 3.82VSHH127 pKa = 6.82NGQVVHH133 pKa = 6.84AAGDD137 pKa = 3.96LIIYY141 pKa = 9.66FGLDD145 pKa = 2.55RR146 pKa = 11.84FANNGDD152 pKa = 3.32AFAGFWFFQDD162 pKa = 3.73NVTLGANGRR171 pKa = 11.84FNGQHH176 pKa = 5.6VARR179 pKa = 11.84QGMEE183 pKa = 3.9GQPGYY188 pKa = 11.31VPGDD192 pKa = 3.42LLVLVEE198 pKa = 4.33YY199 pKa = 9.55PQGANAQPEE208 pKa = 4.43IKK210 pKa = 10.43VYY212 pKa = 10.22EE213 pKa = 4.32WDD215 pKa = 4.47PLDD218 pKa = 5.09LDD220 pKa = 4.53HH221 pKa = 8.09DD222 pKa = 4.33NVAPNLDD229 pKa = 3.39QRR231 pKa = 11.84YY232 pKa = 9.18SSAAAKK238 pKa = 10.44CDD240 pKa = 3.33GSGNKK245 pKa = 8.66LACAITNVANLPGAPEE261 pKa = 3.77WPYY264 pKa = 10.01TPKK267 pKa = 10.8NGFAGIPFEE276 pKa = 4.59SFFEE280 pKa = 4.27GGINVSKK287 pKa = 10.9LLGGDD292 pKa = 3.68VPCFSSFLAEE302 pKa = 4.17TRR304 pKa = 11.84SSRR307 pKa = 11.84SEE309 pKa = 3.78TSQLKK314 pKa = 10.83DD315 pKa = 3.52FVLGDD320 pKa = 3.95FNLCSIAVTKK330 pKa = 10.62QCTAAVDD337 pKa = 3.65AGDD340 pKa = 4.24GGNSVLVNYY349 pKa = 10.51SGTVTNDD356 pKa = 2.7GGLPLFDD363 pKa = 3.5VTVTDD368 pKa = 5.9DD369 pKa = 3.2MGTANTADD377 pKa = 3.63DD378 pKa = 3.95VVVFGPATLQPGEE391 pKa = 4.22SQPYY395 pKa = 7.63NGSYY399 pKa = 8.85STLSIPATDD408 pKa = 3.36EE409 pKa = 3.91VTAVGHH415 pKa = 6.44RR416 pKa = 11.84NGTSVEE422 pKa = 4.05ATAEE426 pKa = 4.34ASCSPDD432 pKa = 3.21VTPALAVDD440 pKa = 4.54KK441 pKa = 11.2ACTANIASGGAGIDD455 pKa = 3.29VLFNGTVTNTGNVALEE471 pKa = 4.0NVMVVDD477 pKa = 5.49DD478 pKa = 5.18GGTPGDD484 pKa = 3.73TSDD487 pKa = 4.36DD488 pKa = 3.86VTVLGPVTLNAGEE501 pKa = 4.17SMPYY505 pKa = 9.87NGSFSASGSNSSTDD519 pKa = 3.15TVTATGSDD527 pKa = 3.85VLTQSPVEE535 pKa = 4.03ATAQATCEE543 pKa = 4.05ADD545 pKa = 3.3VTPAIEE551 pKa = 4.2VTKK554 pKa = 10.67QCSANINTGGTGIDD568 pKa = 3.32VLFTGTVTNTGDD580 pKa = 3.25VALGNVTVVDD590 pKa = 4.43DD591 pKa = 4.55NGTPGDD597 pKa = 3.76TSDD600 pKa = 4.36DD601 pKa = 3.85VTVLGPISLAPGEE614 pKa = 4.26SANYY618 pKa = 9.5NGSFSASGSSSTDD631 pKa = 2.84TVTASGADD639 pKa = 3.59VLTQSPVEE647 pKa = 4.12AQASASCSADD657 pKa = 3.04VTPAIEE663 pKa = 4.21VTKK666 pKa = 10.74QCTDD670 pKa = 2.93APAFGQAILFDD681 pKa = 3.82GTVTNTGNVALLNVTVVDD699 pKa = 4.68DD700 pKa = 4.4NGTPGDD706 pKa = 3.81TSDD709 pKa = 5.61DD710 pKa = 3.62VTFNLGDD717 pKa = 4.13LAPGAAANYY726 pKa = 9.03NGAYY730 pKa = 8.14TPSLAGSYY738 pKa = 9.71TNTVVATAEE747 pKa = 4.19DD748 pKa = 4.14AVEE751 pKa = 4.07SSPVSSSADD760 pKa = 3.35ATCEE764 pKa = 3.99VPPPPEE770 pKa = 5.48GEE772 pKa = 4.16GCTPGFWKK780 pKa = 10.55NSPGSWGPTGYY791 pKa = 10.55SPNQTVGSVFSLPSGQLANQLGGNSLIQALNYY823 pKa = 10.69AGGNNLLGSAQILLRR838 pKa = 11.84AAVAAVLNASHH849 pKa = 7.71PDD851 pKa = 2.81VDD853 pKa = 4.28YY854 pKa = 10.37PRR856 pKa = 11.84TAASVIADD864 pKa = 3.58VNAALATKK872 pKa = 10.43DD873 pKa = 3.17RR874 pKa = 11.84ATILALAAALDD885 pKa = 3.9ADD887 pKa = 4.3NNLGCDD893 pKa = 3.99LPNDD897 pKa = 3.53NSFF900 pKa = 3.53
Molecular weight: 91.28 kDa
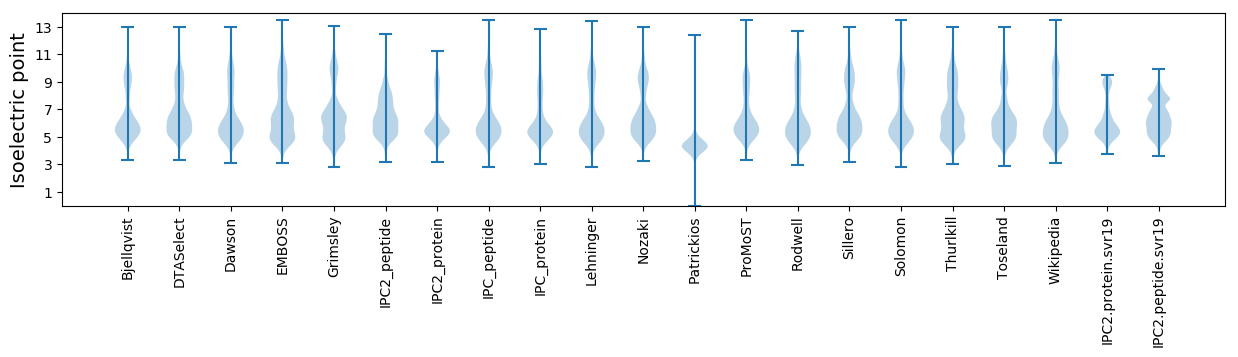
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9LRJ8|A0A1G9LRJ8_9PSED Uncharacterized protein (Fragment) OS=Pseudomonas indica OX=137658 GN=SAMN05216186_1261 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.31RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.31RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1715093 |
25 |
14206 |
331.1 |
36.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.203 ± 0.04 | 1.001 ± 0.013 |
5.534 ± 0.037 | 6.195 ± 0.036 |
3.578 ± 0.022 | 8.152 ± 0.041 |
2.234 ± 0.019 | 4.442 ± 0.024 |
3.128 ± 0.029 | 12.166 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.126 ± 0.018 | 2.806 ± 0.022 |
5.079 ± 0.026 | 4.271 ± 0.025 |
7.168 ± 0.035 | 5.448 ± 0.033 |
4.6 ± 0.029 | 6.889 ± 0.03 |
1.458 ± 0.015 | 2.52 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
