
Halomonas elongata (strain ATCC 33173 / DSM 2581 / NBRC 15536 / NCIMB 2198 / 1H9)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; Halomonas elongata
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
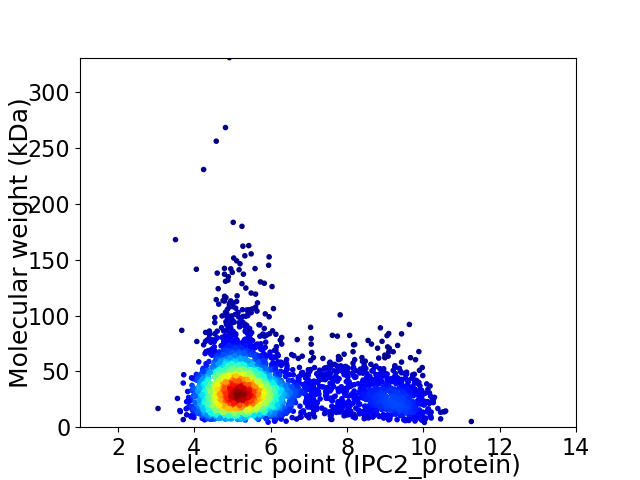
Virtual 2D-PAGE plot for 3622 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1V876|E1V876_HALED Uncharacterized protein OS=Halomonas elongata (strain ATCC 33173 / DSM 2581 / NBRC 15536 / NCIMB 2198 / 1H9) OX=768066 GN=HELO_1755 PE=4 SV=1
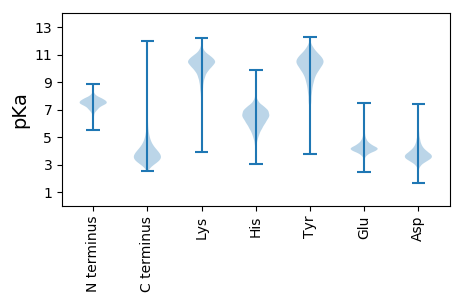
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAMAASGAQAATVYY25 pKa = 10.61NQDD28 pKa = 3.01GTKK31 pKa = 10.39LDD33 pKa = 3.23IYY35 pKa = 11.2GNVQIGFRR43 pKa = 11.84NIEE46 pKa = 3.91AEE48 pKa = 3.87NDD50 pKa = 3.15NGNIEE55 pKa = 4.25TQNDD59 pKa = 3.44VFDD62 pKa = 4.24NGSTIGFAAEE72 pKa = 3.66HH73 pKa = 6.23VIYY76 pKa = 10.73DD77 pKa = 3.83GLTGYY82 pKa = 9.48MKK84 pKa = 10.25IEE86 pKa = 4.19FDD88 pKa = 4.26DD89 pKa = 5.09FKK91 pKa = 11.42ADD93 pKa = 3.42EE94 pKa = 4.51MKK96 pKa = 10.32TAGRR100 pKa = 11.84DD101 pKa = 3.55AGDD104 pKa = 3.3TAYY107 pKa = 10.99VGLKK111 pKa = 10.83GNFGDD116 pKa = 4.31VKK118 pKa = 10.62LGSYY122 pKa = 7.64DD123 pKa = 3.36TLMDD127 pKa = 4.74DD128 pKa = 5.12WIQDD132 pKa = 3.82PITNNEE138 pKa = 3.98YY139 pKa = 10.88FDD141 pKa = 4.29VSDD144 pKa = 3.65TSGSGSSVVAVGGEE158 pKa = 4.17VEE160 pKa = 4.38TDD162 pKa = 2.79QLTYY166 pKa = 10.89VSPSFNGLEE175 pKa = 4.06LAIGTQYY182 pKa = 10.89KK183 pKa = 10.99GDD185 pKa = 3.8MEE187 pKa = 4.5EE188 pKa = 4.38EE189 pKa = 4.39NVTSRR194 pKa = 11.84GNASVFGGAKK204 pKa = 8.05YY205 pKa = 8.26TAGNFSVAATYY216 pKa = 11.17DD217 pKa = 3.3NLDD220 pKa = 3.74NYY222 pKa = 10.59EE223 pKa = 4.08VTQTGVDD230 pKa = 3.42NKK232 pKa = 10.62QEE234 pKa = 3.98FGDD237 pKa = 3.86RR238 pKa = 11.84YY239 pKa = 10.45GVTGQYY245 pKa = 8.27QWNSLRR251 pKa = 11.84VALKK255 pKa = 9.73YY256 pKa = 10.72EE257 pKa = 4.33RR258 pKa = 11.84FDD260 pKa = 3.99SDD262 pKa = 3.81LDD264 pKa = 3.89NVDD267 pKa = 3.35SVNFYY272 pKa = 11.5GLGARR277 pKa = 11.84YY278 pKa = 9.62GYY280 pKa = 10.9GYY282 pKa = 10.72GDD284 pKa = 3.22IYY286 pKa = 11.2GAYY289 pKa = 9.76QYY291 pKa = 11.48VDD293 pKa = 3.13VGGDD297 pKa = 3.23TFGNVVDD304 pKa = 5.22DD305 pKa = 4.25ATSGDD310 pKa = 4.2SPSDD314 pKa = 3.49TASDD318 pKa = 4.38RR319 pKa = 11.84GDD321 pKa = 3.2DD322 pKa = 3.83TYY324 pKa = 11.96NEE326 pKa = 4.44FIIGGTYY333 pKa = 10.26NISDD337 pKa = 3.69AMYY340 pKa = 8.35TWVEE344 pKa = 3.36AAFYY348 pKa = 10.74DD349 pKa = 4.74RR350 pKa = 11.84EE351 pKa = 4.21DD352 pKa = 4.28DD353 pKa = 3.97EE354 pKa = 7.12GDD356 pKa = 3.44GVAAGVTYY364 pKa = 9.67MFF366 pKa = 5.27
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAMAASGAQAATVYY25 pKa = 10.61NQDD28 pKa = 3.01GTKK31 pKa = 10.39LDD33 pKa = 3.23IYY35 pKa = 11.2GNVQIGFRR43 pKa = 11.84NIEE46 pKa = 3.91AEE48 pKa = 3.87NDD50 pKa = 3.15NGNIEE55 pKa = 4.25TQNDD59 pKa = 3.44VFDD62 pKa = 4.24NGSTIGFAAEE72 pKa = 3.66HH73 pKa = 6.23VIYY76 pKa = 10.73DD77 pKa = 3.83GLTGYY82 pKa = 9.48MKK84 pKa = 10.25IEE86 pKa = 4.19FDD88 pKa = 4.26DD89 pKa = 5.09FKK91 pKa = 11.42ADD93 pKa = 3.42EE94 pKa = 4.51MKK96 pKa = 10.32TAGRR100 pKa = 11.84DD101 pKa = 3.55AGDD104 pKa = 3.3TAYY107 pKa = 10.99VGLKK111 pKa = 10.83GNFGDD116 pKa = 4.31VKK118 pKa = 10.62LGSYY122 pKa = 7.64DD123 pKa = 3.36TLMDD127 pKa = 4.74DD128 pKa = 5.12WIQDD132 pKa = 3.82PITNNEE138 pKa = 3.98YY139 pKa = 10.88FDD141 pKa = 4.29VSDD144 pKa = 3.65TSGSGSSVVAVGGEE158 pKa = 4.17VEE160 pKa = 4.38TDD162 pKa = 2.79QLTYY166 pKa = 10.89VSPSFNGLEE175 pKa = 4.06LAIGTQYY182 pKa = 10.89KK183 pKa = 10.99GDD185 pKa = 3.8MEE187 pKa = 4.5EE188 pKa = 4.38EE189 pKa = 4.39NVTSRR194 pKa = 11.84GNASVFGGAKK204 pKa = 8.05YY205 pKa = 8.26TAGNFSVAATYY216 pKa = 11.17DD217 pKa = 3.3NLDD220 pKa = 3.74NYY222 pKa = 10.59EE223 pKa = 4.08VTQTGVDD230 pKa = 3.42NKK232 pKa = 10.62QEE234 pKa = 3.98FGDD237 pKa = 3.86RR238 pKa = 11.84YY239 pKa = 10.45GVTGQYY245 pKa = 8.27QWNSLRR251 pKa = 11.84VALKK255 pKa = 9.73YY256 pKa = 10.72EE257 pKa = 4.33RR258 pKa = 11.84FDD260 pKa = 3.99SDD262 pKa = 3.81LDD264 pKa = 3.89NVDD267 pKa = 3.35SVNFYY272 pKa = 11.5GLGARR277 pKa = 11.84YY278 pKa = 9.62GYY280 pKa = 10.9GYY282 pKa = 10.72GDD284 pKa = 3.22IYY286 pKa = 11.2GAYY289 pKa = 9.76QYY291 pKa = 11.48VDD293 pKa = 3.13VGGDD297 pKa = 3.23TFGNVVDD304 pKa = 5.22DD305 pKa = 4.25ATSGDD310 pKa = 4.2SPSDD314 pKa = 3.49TASDD318 pKa = 4.38RR319 pKa = 11.84GDD321 pKa = 3.2DD322 pKa = 3.83TYY324 pKa = 11.96NEE326 pKa = 4.44FIIGGTYY333 pKa = 10.26NISDD337 pKa = 3.69AMYY340 pKa = 8.35TWVEE344 pKa = 3.36AAFYY348 pKa = 10.74DD349 pKa = 4.74RR350 pKa = 11.84EE351 pKa = 4.21DD352 pKa = 4.28DD353 pKa = 3.97EE354 pKa = 7.12GDD356 pKa = 3.44GVAAGVTYY364 pKa = 9.67MFF366 pKa = 5.27
Molecular weight: 39.57 kDa
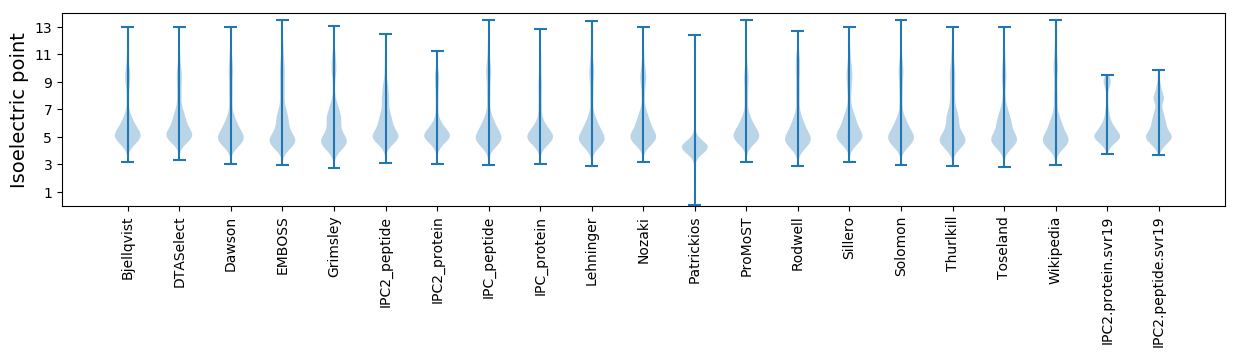
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1VCK1|E1VCK1_HALED DUF461 family protein OS=Halomonas elongata (strain ATCC 33173 / DSM 2581 / NBRC 15536 / NCIMB 2198 / 1H9) OX=768066 GN=HELO_2472 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1202401 |
37 |
3024 |
332.0 |
36.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.184 ± 0.043 | 0.914 ± 0.012 |
6.09 ± 0.042 | 6.72 ± 0.05 |
3.383 ± 0.026 | 8.367 ± 0.037 |
2.47 ± 0.019 | 4.542 ± 0.036 |
2.439 ± 0.029 | 11.382 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.515 ± 0.019 | 2.437 ± 0.021 |
4.984 ± 0.028 | 3.614 ± 0.03 |
7.562 ± 0.046 | 5.455 ± 0.028 |
4.988 ± 0.023 | 7.137 ± 0.034 |
1.503 ± 0.019 | 2.312 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
