
Hubei virga-like virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
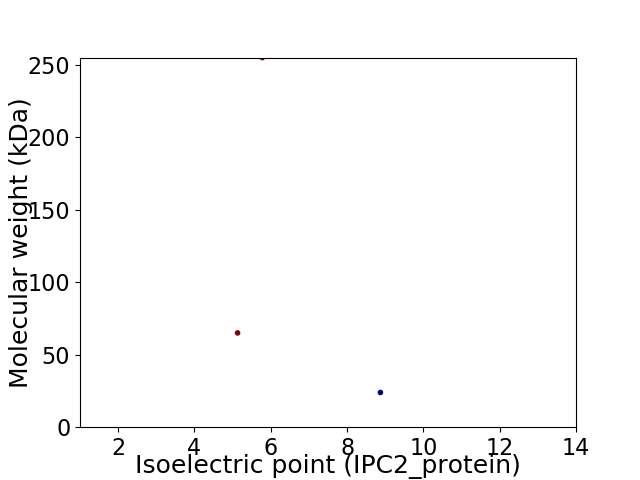
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
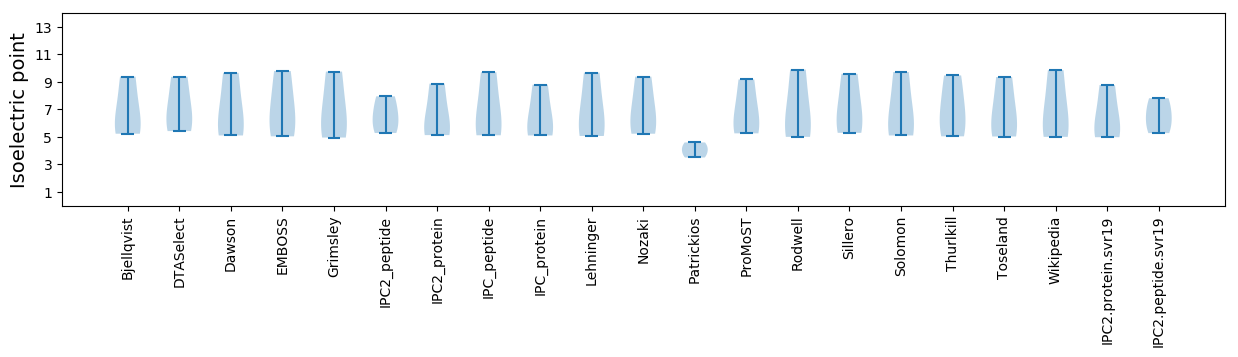
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJW0|A0A1L3KJW0_9VIRU Uncharacterized protein OS=Hubei virga-like virus 17 OX=1923332 PE=4 SV=1
MM1 pKa = 7.6LKK3 pKa = 10.52FILAEE8 pKa = 3.93FEE10 pKa = 4.12NFAKK14 pKa = 10.86VKK16 pKa = 8.87FTMFRR21 pKa = 11.84LVLILIIEE29 pKa = 4.37VVVVNCKK36 pKa = 9.67NYY38 pKa = 9.32RR39 pKa = 11.84QEE41 pKa = 4.09VVINNVSLVKK51 pKa = 10.49LDD53 pKa = 4.21LVPLFAFEE61 pKa = 4.38TGVPVYY67 pKa = 10.63FKK69 pKa = 11.21DD70 pKa = 5.38DD71 pKa = 4.0LDD73 pKa = 4.28LKK75 pKa = 10.54NLLVMRR81 pKa = 11.84EE82 pKa = 3.9LDD84 pKa = 3.62YY85 pKa = 11.63LKK87 pKa = 11.03NFYY90 pKa = 10.64TEE92 pKa = 3.78MNNLYY97 pKa = 10.34RR98 pKa = 11.84YY99 pKa = 9.87FEE101 pKa = 4.25YY102 pKa = 10.84NYY104 pKa = 10.18VRR106 pKa = 11.84VKK108 pKa = 9.46LHH110 pKa = 6.6CLDD113 pKa = 4.3ALSEE117 pKa = 4.11NLIQDD122 pKa = 3.79YY123 pKa = 10.82DD124 pKa = 4.11CEE126 pKa = 4.49LDD128 pKa = 4.02YY129 pKa = 11.51SCEE132 pKa = 3.52PHH134 pKa = 7.37IRR136 pKa = 11.84IKK138 pKa = 10.54VAHH141 pKa = 5.74TWYY144 pKa = 10.62GSAVYY149 pKa = 10.35ACLGLNRR156 pKa = 11.84TRR158 pKa = 11.84VFNSTNVLVRR168 pKa = 11.84VHH170 pKa = 6.74KK171 pKa = 10.54LAFPGVGIYY180 pKa = 8.51MKK182 pKa = 10.29NVSDD186 pKa = 4.43IYY188 pKa = 11.57GNLDD192 pKa = 3.66DD193 pKa = 5.72SGLRR197 pKa = 11.84YY198 pKa = 10.15DD199 pKa = 5.08DD200 pKa = 5.69LVLADD205 pKa = 3.93YY206 pKa = 10.81EE207 pKa = 4.19IDD209 pKa = 3.56GVRR212 pKa = 11.84KK213 pKa = 9.69FIFTYY218 pKa = 10.33KK219 pKa = 10.82YY220 pKa = 9.94MLNQSSAAFTDD231 pKa = 4.3AYY233 pKa = 10.35RR234 pKa = 11.84IYY236 pKa = 10.97GDD238 pKa = 3.32WDD240 pKa = 3.66NVCVEE245 pKa = 4.23NKK247 pKa = 9.32VSKK250 pKa = 10.46FMYY253 pKa = 10.24YY254 pKa = 10.03YY255 pKa = 10.21FGYY258 pKa = 8.56IAKK261 pKa = 10.59YY262 pKa = 9.94NGTDD266 pKa = 2.94KK267 pKa = 11.03VIPHH271 pKa = 6.59MPLMVYY277 pKa = 9.89VGRR280 pKa = 11.84RR281 pKa = 11.84NAAADD286 pKa = 3.61YY287 pKa = 10.3ASEE290 pKa = 4.01FFTYY294 pKa = 8.36YY295 pKa = 10.33QCTEE299 pKa = 4.09EE300 pKa = 4.25NLKK303 pKa = 10.74EE304 pKa = 3.9PLLVNVGGLDD314 pKa = 3.35FLKK317 pKa = 10.81YY318 pKa = 10.57NKK320 pKa = 9.33PNCTDD325 pKa = 3.57YY326 pKa = 11.74VKK328 pKa = 10.88VSSDD332 pKa = 3.22EE333 pKa = 4.14FQCNVIFDD341 pKa = 4.63LGNPFLKK348 pKa = 10.61AICPDD353 pKa = 3.64DD354 pKa = 4.86YY355 pKa = 11.04IVKK358 pKa = 10.26KK359 pKa = 10.86DD360 pKa = 3.77VVDD363 pKa = 3.83HH364 pKa = 6.53NKK366 pKa = 10.07NDD368 pKa = 3.26FDD370 pKa = 4.91TEE372 pKa = 4.48VNCIKK377 pKa = 10.49VRR379 pKa = 11.84TIKK382 pKa = 10.68KK383 pKa = 9.0SWFSEE388 pKa = 3.87LFVFLEE394 pKa = 4.12KK395 pKa = 10.51KK396 pKa = 10.06VLHH399 pKa = 5.89LVEE402 pKa = 5.67GIIKK406 pKa = 10.18EE407 pKa = 4.0IAQVLSEE414 pKa = 4.31SVKK417 pKa = 10.26VLVDD421 pKa = 3.46EE422 pKa = 4.41LVKK425 pKa = 10.68VLKK428 pKa = 11.02DD429 pKa = 3.06MGPIFKK435 pKa = 9.96PLLEE439 pKa = 6.0DD440 pKa = 2.85IFKK443 pKa = 10.52DD444 pKa = 3.38VKK446 pKa = 10.67EE447 pKa = 3.88IAKK450 pKa = 10.14EE451 pKa = 3.98VFKK454 pKa = 11.32DD455 pKa = 4.01FGQIFKK461 pKa = 10.77DD462 pKa = 3.32IFADD466 pKa = 3.96AVEE469 pKa = 4.61LLDD472 pKa = 5.11DD473 pKa = 4.81LVNFTLGFVFGIIRR487 pKa = 11.84YY488 pKa = 8.54LLNLLVQLEE497 pKa = 3.81KK498 pKa = 10.62DD499 pKa = 3.8YY500 pKa = 11.6YY501 pKa = 10.09VFEE504 pKa = 4.24ILVAYY509 pKa = 9.57VVLRR513 pKa = 11.84ILLVHH518 pKa = 6.27NLSVVVILVIVCLTFGVEE536 pKa = 3.57RR537 pKa = 11.84RR538 pKa = 11.84YY539 pKa = 11.26DD540 pKa = 3.65SVVYY544 pKa = 9.63DD545 pKa = 3.36IWSNVTIDD553 pKa = 3.73MKK555 pKa = 11.24LLNN558 pKa = 4.37
MM1 pKa = 7.6LKK3 pKa = 10.52FILAEE8 pKa = 3.93FEE10 pKa = 4.12NFAKK14 pKa = 10.86VKK16 pKa = 8.87FTMFRR21 pKa = 11.84LVLILIIEE29 pKa = 4.37VVVVNCKK36 pKa = 9.67NYY38 pKa = 9.32RR39 pKa = 11.84QEE41 pKa = 4.09VVINNVSLVKK51 pKa = 10.49LDD53 pKa = 4.21LVPLFAFEE61 pKa = 4.38TGVPVYY67 pKa = 10.63FKK69 pKa = 11.21DD70 pKa = 5.38DD71 pKa = 4.0LDD73 pKa = 4.28LKK75 pKa = 10.54NLLVMRR81 pKa = 11.84EE82 pKa = 3.9LDD84 pKa = 3.62YY85 pKa = 11.63LKK87 pKa = 11.03NFYY90 pKa = 10.64TEE92 pKa = 3.78MNNLYY97 pKa = 10.34RR98 pKa = 11.84YY99 pKa = 9.87FEE101 pKa = 4.25YY102 pKa = 10.84NYY104 pKa = 10.18VRR106 pKa = 11.84VKK108 pKa = 9.46LHH110 pKa = 6.6CLDD113 pKa = 4.3ALSEE117 pKa = 4.11NLIQDD122 pKa = 3.79YY123 pKa = 10.82DD124 pKa = 4.11CEE126 pKa = 4.49LDD128 pKa = 4.02YY129 pKa = 11.51SCEE132 pKa = 3.52PHH134 pKa = 7.37IRR136 pKa = 11.84IKK138 pKa = 10.54VAHH141 pKa = 5.74TWYY144 pKa = 10.62GSAVYY149 pKa = 10.35ACLGLNRR156 pKa = 11.84TRR158 pKa = 11.84VFNSTNVLVRR168 pKa = 11.84VHH170 pKa = 6.74KK171 pKa = 10.54LAFPGVGIYY180 pKa = 8.51MKK182 pKa = 10.29NVSDD186 pKa = 4.43IYY188 pKa = 11.57GNLDD192 pKa = 3.66DD193 pKa = 5.72SGLRR197 pKa = 11.84YY198 pKa = 10.15DD199 pKa = 5.08DD200 pKa = 5.69LVLADD205 pKa = 3.93YY206 pKa = 10.81EE207 pKa = 4.19IDD209 pKa = 3.56GVRR212 pKa = 11.84KK213 pKa = 9.69FIFTYY218 pKa = 10.33KK219 pKa = 10.82YY220 pKa = 9.94MLNQSSAAFTDD231 pKa = 4.3AYY233 pKa = 10.35RR234 pKa = 11.84IYY236 pKa = 10.97GDD238 pKa = 3.32WDD240 pKa = 3.66NVCVEE245 pKa = 4.23NKK247 pKa = 9.32VSKK250 pKa = 10.46FMYY253 pKa = 10.24YY254 pKa = 10.03YY255 pKa = 10.21FGYY258 pKa = 8.56IAKK261 pKa = 10.59YY262 pKa = 9.94NGTDD266 pKa = 2.94KK267 pKa = 11.03VIPHH271 pKa = 6.59MPLMVYY277 pKa = 9.89VGRR280 pKa = 11.84RR281 pKa = 11.84NAAADD286 pKa = 3.61YY287 pKa = 10.3ASEE290 pKa = 4.01FFTYY294 pKa = 8.36YY295 pKa = 10.33QCTEE299 pKa = 4.09EE300 pKa = 4.25NLKK303 pKa = 10.74EE304 pKa = 3.9PLLVNVGGLDD314 pKa = 3.35FLKK317 pKa = 10.81YY318 pKa = 10.57NKK320 pKa = 9.33PNCTDD325 pKa = 3.57YY326 pKa = 11.74VKK328 pKa = 10.88VSSDD332 pKa = 3.22EE333 pKa = 4.14FQCNVIFDD341 pKa = 4.63LGNPFLKK348 pKa = 10.61AICPDD353 pKa = 3.64DD354 pKa = 4.86YY355 pKa = 11.04IVKK358 pKa = 10.26KK359 pKa = 10.86DD360 pKa = 3.77VVDD363 pKa = 3.83HH364 pKa = 6.53NKK366 pKa = 10.07NDD368 pKa = 3.26FDD370 pKa = 4.91TEE372 pKa = 4.48VNCIKK377 pKa = 10.49VRR379 pKa = 11.84TIKK382 pKa = 10.68KK383 pKa = 9.0SWFSEE388 pKa = 3.87LFVFLEE394 pKa = 4.12KK395 pKa = 10.51KK396 pKa = 10.06VLHH399 pKa = 5.89LVEE402 pKa = 5.67GIIKK406 pKa = 10.18EE407 pKa = 4.0IAQVLSEE414 pKa = 4.31SVKK417 pKa = 10.26VLVDD421 pKa = 3.46EE422 pKa = 4.41LVKK425 pKa = 10.68VLKK428 pKa = 11.02DD429 pKa = 3.06MGPIFKK435 pKa = 9.96PLLEE439 pKa = 6.0DD440 pKa = 2.85IFKK443 pKa = 10.52DD444 pKa = 3.38VKK446 pKa = 10.67EE447 pKa = 3.88IAKK450 pKa = 10.14EE451 pKa = 3.98VFKK454 pKa = 11.32DD455 pKa = 4.01FGQIFKK461 pKa = 10.77DD462 pKa = 3.32IFADD466 pKa = 3.96AVEE469 pKa = 4.61LLDD472 pKa = 5.11DD473 pKa = 4.81LVNFTLGFVFGIIRR487 pKa = 11.84YY488 pKa = 8.54LLNLLVQLEE497 pKa = 3.81KK498 pKa = 10.62DD499 pKa = 3.8YY500 pKa = 11.6YY501 pKa = 10.09VFEE504 pKa = 4.24ILVAYY509 pKa = 9.57VVLRR513 pKa = 11.84ILLVHH518 pKa = 6.27NLSVVVILVIVCLTFGVEE536 pKa = 3.57RR537 pKa = 11.84RR538 pKa = 11.84YY539 pKa = 11.26DD540 pKa = 3.65SVVYY544 pKa = 9.63DD545 pKa = 3.36IWSNVTIDD553 pKa = 3.73MKK555 pKa = 11.24LLNN558 pKa = 4.37
Molecular weight: 65.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJW0|A0A1L3KJW0_9VIRU Uncharacterized protein OS=Hubei virga-like virus 17 OX=1923332 PE=4 SV=1
MM1 pKa = 7.5EE2 pKa = 5.3AQPQQMQGNGRR13 pKa = 11.84TGWRR17 pKa = 11.84VEE19 pKa = 4.14NKK21 pKa = 10.11VGDD24 pKa = 4.08LDD26 pKa = 4.76IISDD30 pKa = 3.61VLGAYY35 pKa = 8.81KK36 pKa = 10.28RR37 pKa = 11.84LPMYY41 pKa = 7.7PFPMFVLMLTLIVGLAEE58 pKa = 4.28IYY60 pKa = 10.84NGGEE64 pKa = 3.85KK65 pKa = 10.04PLEE68 pKa = 3.87YY69 pKa = 10.54LLNGIQADD77 pKa = 4.23LNNEE81 pKa = 4.47NIPTWEE87 pKa = 3.9KK88 pKa = 9.69TALKK92 pKa = 8.9ICKK95 pKa = 9.26YY96 pKa = 9.25VLEE99 pKa = 5.14FIIAHH104 pKa = 6.66KK105 pKa = 10.36IKK107 pKa = 10.91VFGFLMISIPVIIRR121 pKa = 11.84PSKK124 pKa = 8.12NNVYY128 pKa = 9.93IWVLLVAILIMMRR141 pKa = 11.84NWGFFEE147 pKa = 4.76YY148 pKa = 10.22IVIGNLFYY156 pKa = 10.42IYY158 pKa = 9.72TQLLSLTNKK167 pKa = 9.76FLIAMLIVVVVLWNFVLPVKK187 pKa = 10.37SNKK190 pKa = 9.28IPPGAPSPPPVVSGPAPPSRR210 pKa = 11.84KK211 pKa = 7.22TT212 pKa = 3.24
MM1 pKa = 7.5EE2 pKa = 5.3AQPQQMQGNGRR13 pKa = 11.84TGWRR17 pKa = 11.84VEE19 pKa = 4.14NKK21 pKa = 10.11VGDD24 pKa = 4.08LDD26 pKa = 4.76IISDD30 pKa = 3.61VLGAYY35 pKa = 8.81KK36 pKa = 10.28RR37 pKa = 11.84LPMYY41 pKa = 7.7PFPMFVLMLTLIVGLAEE58 pKa = 4.28IYY60 pKa = 10.84NGGEE64 pKa = 3.85KK65 pKa = 10.04PLEE68 pKa = 3.87YY69 pKa = 10.54LLNGIQADD77 pKa = 4.23LNNEE81 pKa = 4.47NIPTWEE87 pKa = 3.9KK88 pKa = 9.69TALKK92 pKa = 8.9ICKK95 pKa = 9.26YY96 pKa = 9.25VLEE99 pKa = 5.14FIIAHH104 pKa = 6.66KK105 pKa = 10.36IKK107 pKa = 10.91VFGFLMISIPVIIRR121 pKa = 11.84PSKK124 pKa = 8.12NNVYY128 pKa = 9.93IWVLLVAILIMMRR141 pKa = 11.84NWGFFEE147 pKa = 4.76YY148 pKa = 10.22IVIGNLFYY156 pKa = 10.42IYY158 pKa = 9.72TQLLSLTNKK167 pKa = 9.76FLIAMLIVVVVLWNFVLPVKK187 pKa = 10.37SNKK190 pKa = 9.28IPPGAPSPPPVVSGPAPPSRR210 pKa = 11.84KK211 pKa = 7.22TT212 pKa = 3.24
Molecular weight: 23.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3009 |
212 |
2239 |
1003.0 |
114.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.018 ± 0.378 | 2.127 ± 0.443 |
7.511 ± 1.488 | 5.716 ± 0.394 |
5.45 ± 0.526 | 5.218 ± 0.608 |
1.961 ± 0.49 | 5.849 ± 1.417 |
8.109 ± 0.588 | 9.837 ± 1.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.452 | 5.018 ± 0.719 |
3.091 ± 1.418 | 2.193 ± 0.309 |
4.021 ± 0.442 | 6.181 ± 1.323 |
4.021 ± 0.465 | 9.837 ± 1.113 |
0.931 ± 0.375 | 5.018 ± 0.687 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
