
Heterobasidion partitivirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1AB82|E1AB82_9VIRU Putative coat protein OS=Heterobasidion partitivirus 2 OX=872291 PE=4 SV=1
MM1 pKa = 7.05STNPPEE7 pKa = 4.27VLLPLPEE14 pKa = 4.2VDD16 pKa = 3.79PNANNVRR23 pKa = 11.84YY24 pKa = 9.87AKK26 pKa = 10.9LLDD29 pKa = 3.52QYY31 pKa = 10.96RR32 pKa = 11.84ANPSNSNQKK41 pKa = 10.44LLLEE45 pKa = 4.36YY46 pKa = 10.87AEE48 pKa = 4.0QHH50 pKa = 5.48GFNYY54 pKa = 9.08FIPTDD59 pKa = 3.52VPLPDD64 pKa = 4.49DD65 pKa = 4.2RR66 pKa = 11.84KK67 pKa = 9.51SAPGILSLDD76 pKa = 3.52GFRR79 pKa = 11.84FHH81 pKa = 6.47TVAAFHH87 pKa = 7.4RR88 pKa = 11.84YY89 pKa = 7.54QKK91 pKa = 9.8MSRR94 pKa = 11.84YY95 pKa = 9.59GYY97 pKa = 9.62NALGFIFKK105 pKa = 10.89LILTLFNPFLWILTDD120 pKa = 3.18YY121 pKa = 10.55CRR123 pKa = 11.84PSGSADD129 pKa = 2.93AVFEE133 pKa = 4.13NFNQEE138 pKa = 4.02VSPVEE143 pKa = 3.91HH144 pKa = 6.44VNPSRR149 pKa = 11.84LAQIMPLIHH158 pKa = 6.79HH159 pKa = 6.94FFAIKK164 pKa = 9.36PFCPIAFPDD173 pKa = 3.33LRR175 pKa = 11.84FYY177 pKa = 10.81KK178 pKa = 10.08WSLVTSADD186 pKa = 3.51YY187 pKa = 10.17HH188 pKa = 5.98AHH190 pKa = 6.38HH191 pKa = 7.1SKK193 pKa = 10.68DD194 pKa = 3.55QQDD197 pKa = 3.47EE198 pKa = 4.3SAHH201 pKa = 4.8YY202 pKa = 8.19WKK204 pKa = 10.36HH205 pKa = 6.37LKK207 pKa = 10.72DD208 pKa = 3.72SDD210 pKa = 4.06LLQDD214 pKa = 3.5RR215 pKa = 11.84FDD217 pKa = 3.83YY218 pKa = 10.92SDD220 pKa = 3.4RR221 pKa = 11.84PRR223 pKa = 11.84SKK225 pKa = 10.57GYY227 pKa = 10.26FFNTVLLSTRR237 pKa = 11.84TIVHH241 pKa = 6.64NIKK244 pKa = 9.99YY245 pKa = 9.83HH246 pKa = 5.78CLPFQRR252 pKa = 11.84HH253 pKa = 5.17KK254 pKa = 10.9RR255 pKa = 11.84DD256 pKa = 3.27TDD258 pKa = 3.64SSVLQKK264 pKa = 11.0LSFWFMKK271 pKa = 10.1YY272 pKa = 7.74PTVMYY277 pKa = 10.34VRR279 pKa = 11.84SQISKK284 pKa = 9.79LSKK287 pKa = 9.66LKK289 pKa = 10.44VRR291 pKa = 11.84PVYY294 pKa = 9.92NAPFLFILIEE304 pKa = 4.2AMLTLALMAQCRR316 pKa = 11.84LPDD319 pKa = 3.58SCLMWGFEE327 pKa = 4.28TVRR330 pKa = 11.84GGMQEE335 pKa = 4.17LNRR338 pKa = 11.84ISYY341 pKa = 10.69NYY343 pKa = 8.84DD344 pKa = 3.17TFIMIDD350 pKa = 3.26WSRR353 pKa = 11.84YY354 pKa = 8.48DD355 pKa = 3.21QLLPFAIIYY364 pKa = 7.86HH365 pKa = 6.0FWCTFLPQLIRR376 pKa = 11.84VDD378 pKa = 4.09LGYY381 pKa = 10.01MPTEE385 pKa = 4.17QYY387 pKa = 9.11TATQHH392 pKa = 5.42KK393 pKa = 9.52HH394 pKa = 5.51AFTEE398 pKa = 3.87KK399 pKa = 10.83HH400 pKa = 5.81NDD402 pKa = 3.36QKK404 pKa = 11.4EE405 pKa = 4.49SNPEE409 pKa = 3.69YY410 pKa = 10.34ATFASRR416 pKa = 11.84LKK418 pKa = 8.86THH420 pKa = 7.07APHH423 pKa = 7.38IVMFSFIIFNLLAFIWLWYY442 pKa = 9.44VKK444 pKa = 9.62MVFVTPDD451 pKa = 2.61GFGYY455 pKa = 10.45VRR457 pKa = 11.84LLAGVPSGIFMTQICDD473 pKa = 3.68SFCNAFLLIDD483 pKa = 3.81AMLEE487 pKa = 4.12FGFTPDD493 pKa = 4.12DD494 pKa = 3.39IKK496 pKa = 10.87LIRR499 pKa = 11.84MFIQGDD505 pKa = 3.96DD506 pKa = 3.4NVIFYY511 pKa = 10.84LGDD514 pKa = 3.4FTRR517 pKa = 11.84IFAFYY522 pKa = 9.59EE523 pKa = 3.88WLPEE527 pKa = 3.8YY528 pKa = 10.31CQQRR532 pKa = 11.84WHH534 pKa = 5.54MTISVDD540 pKa = 3.08KK541 pKa = 11.24SSITRR546 pKa = 11.84LRR548 pKa = 11.84NKK550 pKa = 10.0IEE552 pKa = 3.74VLGYY556 pKa = 9.4TNLNGMPHH564 pKa = 7.08RR565 pKa = 11.84DD566 pKa = 3.66CAKK569 pKa = 10.73LIATLAYY576 pKa = 9.14PEE578 pKa = 5.64RR579 pKa = 11.84YY580 pKa = 9.46VQDD583 pKa = 3.17KK584 pKa = 9.37TKK586 pKa = 10.91YY587 pKa = 8.47IVFMSRR593 pKa = 11.84AIGIAYY599 pKa = 9.92ANAGHH604 pKa = 6.99DD605 pKa = 3.68RR606 pKa = 11.84QVHH609 pKa = 5.51DD610 pKa = 4.3LCQRR614 pKa = 11.84AYY616 pKa = 10.42LQARR620 pKa = 11.84KK621 pKa = 10.04DD622 pKa = 3.42SGLSYY627 pKa = 11.39DD628 pKa = 3.55EE629 pKa = 4.82LKK631 pKa = 10.86NIKK634 pKa = 9.56IEE636 pKa = 3.99YY637 pKa = 8.7QKK639 pKa = 11.25LGFYY643 pKa = 10.4EE644 pKa = 3.94IFSVNIEE651 pKa = 4.11EE652 pKa = 4.59LRR654 pKa = 11.84EE655 pKa = 3.85HH656 pKa = 7.53LIQDD660 pKa = 3.29VSEE663 pKa = 4.6FPNFHH668 pKa = 7.79DD669 pKa = 3.4IRR671 pKa = 11.84DD672 pKa = 4.13NLRR675 pKa = 11.84HH676 pKa = 4.53WHH678 pKa = 6.38GPHH681 pKa = 5.59TVYY684 pKa = 10.85PMWPRR689 pKa = 11.84HH690 pKa = 5.63FDD692 pKa = 3.85DD693 pKa = 5.4DD694 pKa = 4.4LSSIKK699 pKa = 10.47SPHH702 pKa = 6.52SLTTLYY708 pKa = 11.14DD709 pKa = 3.56VMQSGGLTFDD719 pKa = 3.9YY720 pKa = 11.28NFF722 pKa = 3.99
MM1 pKa = 7.05STNPPEE7 pKa = 4.27VLLPLPEE14 pKa = 4.2VDD16 pKa = 3.79PNANNVRR23 pKa = 11.84YY24 pKa = 9.87AKK26 pKa = 10.9LLDD29 pKa = 3.52QYY31 pKa = 10.96RR32 pKa = 11.84ANPSNSNQKK41 pKa = 10.44LLLEE45 pKa = 4.36YY46 pKa = 10.87AEE48 pKa = 4.0QHH50 pKa = 5.48GFNYY54 pKa = 9.08FIPTDD59 pKa = 3.52VPLPDD64 pKa = 4.49DD65 pKa = 4.2RR66 pKa = 11.84KK67 pKa = 9.51SAPGILSLDD76 pKa = 3.52GFRR79 pKa = 11.84FHH81 pKa = 6.47TVAAFHH87 pKa = 7.4RR88 pKa = 11.84YY89 pKa = 7.54QKK91 pKa = 9.8MSRR94 pKa = 11.84YY95 pKa = 9.59GYY97 pKa = 9.62NALGFIFKK105 pKa = 10.89LILTLFNPFLWILTDD120 pKa = 3.18YY121 pKa = 10.55CRR123 pKa = 11.84PSGSADD129 pKa = 2.93AVFEE133 pKa = 4.13NFNQEE138 pKa = 4.02VSPVEE143 pKa = 3.91HH144 pKa = 6.44VNPSRR149 pKa = 11.84LAQIMPLIHH158 pKa = 6.79HH159 pKa = 6.94FFAIKK164 pKa = 9.36PFCPIAFPDD173 pKa = 3.33LRR175 pKa = 11.84FYY177 pKa = 10.81KK178 pKa = 10.08WSLVTSADD186 pKa = 3.51YY187 pKa = 10.17HH188 pKa = 5.98AHH190 pKa = 6.38HH191 pKa = 7.1SKK193 pKa = 10.68DD194 pKa = 3.55QQDD197 pKa = 3.47EE198 pKa = 4.3SAHH201 pKa = 4.8YY202 pKa = 8.19WKK204 pKa = 10.36HH205 pKa = 6.37LKK207 pKa = 10.72DD208 pKa = 3.72SDD210 pKa = 4.06LLQDD214 pKa = 3.5RR215 pKa = 11.84FDD217 pKa = 3.83YY218 pKa = 10.92SDD220 pKa = 3.4RR221 pKa = 11.84PRR223 pKa = 11.84SKK225 pKa = 10.57GYY227 pKa = 10.26FFNTVLLSTRR237 pKa = 11.84TIVHH241 pKa = 6.64NIKK244 pKa = 9.99YY245 pKa = 9.83HH246 pKa = 5.78CLPFQRR252 pKa = 11.84HH253 pKa = 5.17KK254 pKa = 10.9RR255 pKa = 11.84DD256 pKa = 3.27TDD258 pKa = 3.64SSVLQKK264 pKa = 11.0LSFWFMKK271 pKa = 10.1YY272 pKa = 7.74PTVMYY277 pKa = 10.34VRR279 pKa = 11.84SQISKK284 pKa = 9.79LSKK287 pKa = 9.66LKK289 pKa = 10.44VRR291 pKa = 11.84PVYY294 pKa = 9.92NAPFLFILIEE304 pKa = 4.2AMLTLALMAQCRR316 pKa = 11.84LPDD319 pKa = 3.58SCLMWGFEE327 pKa = 4.28TVRR330 pKa = 11.84GGMQEE335 pKa = 4.17LNRR338 pKa = 11.84ISYY341 pKa = 10.69NYY343 pKa = 8.84DD344 pKa = 3.17TFIMIDD350 pKa = 3.26WSRR353 pKa = 11.84YY354 pKa = 8.48DD355 pKa = 3.21QLLPFAIIYY364 pKa = 7.86HH365 pKa = 6.0FWCTFLPQLIRR376 pKa = 11.84VDD378 pKa = 4.09LGYY381 pKa = 10.01MPTEE385 pKa = 4.17QYY387 pKa = 9.11TATQHH392 pKa = 5.42KK393 pKa = 9.52HH394 pKa = 5.51AFTEE398 pKa = 3.87KK399 pKa = 10.83HH400 pKa = 5.81NDD402 pKa = 3.36QKK404 pKa = 11.4EE405 pKa = 4.49SNPEE409 pKa = 3.69YY410 pKa = 10.34ATFASRR416 pKa = 11.84LKK418 pKa = 8.86THH420 pKa = 7.07APHH423 pKa = 7.38IVMFSFIIFNLLAFIWLWYY442 pKa = 9.44VKK444 pKa = 9.62MVFVTPDD451 pKa = 2.61GFGYY455 pKa = 10.45VRR457 pKa = 11.84LLAGVPSGIFMTQICDD473 pKa = 3.68SFCNAFLLIDD483 pKa = 3.81AMLEE487 pKa = 4.12FGFTPDD493 pKa = 4.12DD494 pKa = 3.39IKK496 pKa = 10.87LIRR499 pKa = 11.84MFIQGDD505 pKa = 3.96DD506 pKa = 3.4NVIFYY511 pKa = 10.84LGDD514 pKa = 3.4FTRR517 pKa = 11.84IFAFYY522 pKa = 9.59EE523 pKa = 3.88WLPEE527 pKa = 3.8YY528 pKa = 10.31CQQRR532 pKa = 11.84WHH534 pKa = 5.54MTISVDD540 pKa = 3.08KK541 pKa = 11.24SSITRR546 pKa = 11.84LRR548 pKa = 11.84NKK550 pKa = 10.0IEE552 pKa = 3.74VLGYY556 pKa = 9.4TNLNGMPHH564 pKa = 7.08RR565 pKa = 11.84DD566 pKa = 3.66CAKK569 pKa = 10.73LIATLAYY576 pKa = 9.14PEE578 pKa = 5.64RR579 pKa = 11.84YY580 pKa = 9.46VQDD583 pKa = 3.17KK584 pKa = 9.37TKK586 pKa = 10.91YY587 pKa = 8.47IVFMSRR593 pKa = 11.84AIGIAYY599 pKa = 9.92ANAGHH604 pKa = 6.99DD605 pKa = 3.68RR606 pKa = 11.84QVHH609 pKa = 5.51DD610 pKa = 4.3LCQRR614 pKa = 11.84AYY616 pKa = 10.42LQARR620 pKa = 11.84KK621 pKa = 10.04DD622 pKa = 3.42SGLSYY627 pKa = 11.39DD628 pKa = 3.55EE629 pKa = 4.82LKK631 pKa = 10.86NIKK634 pKa = 9.56IEE636 pKa = 3.99YY637 pKa = 8.7QKK639 pKa = 11.25LGFYY643 pKa = 10.4EE644 pKa = 3.94IFSVNIEE651 pKa = 4.11EE652 pKa = 4.59LRR654 pKa = 11.84EE655 pKa = 3.85HH656 pKa = 7.53LIQDD660 pKa = 3.29VSEE663 pKa = 4.6FPNFHH668 pKa = 7.79DD669 pKa = 3.4IRR671 pKa = 11.84DD672 pKa = 4.13NLRR675 pKa = 11.84HH676 pKa = 4.53WHH678 pKa = 6.38GPHH681 pKa = 5.59TVYY684 pKa = 10.85PMWPRR689 pKa = 11.84HH690 pKa = 5.63FDD692 pKa = 3.85DD693 pKa = 5.4DD694 pKa = 4.4LSSIKK699 pKa = 10.47SPHH702 pKa = 6.52SLTTLYY708 pKa = 11.14DD709 pKa = 3.56VMQSGGLTFDD719 pKa = 3.9YY720 pKa = 11.28NFF722 pKa = 3.99
Molecular weight: 84.88 kDa
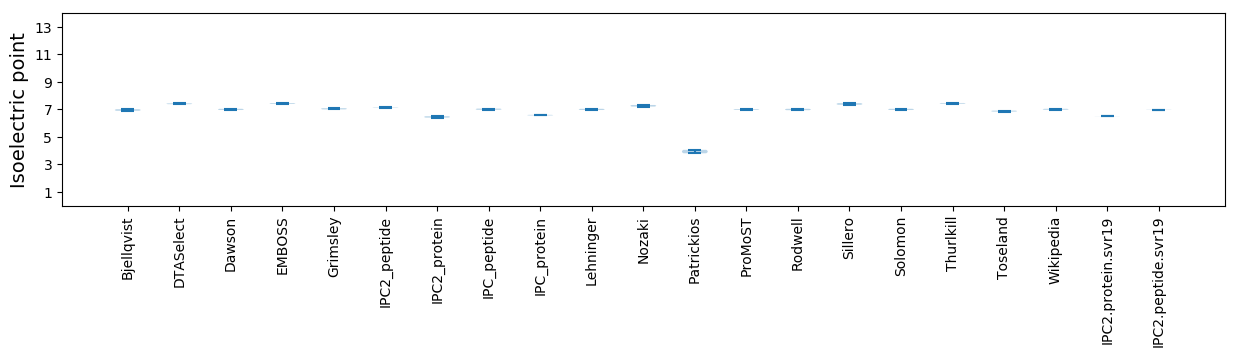
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1AB82|E1AB82_9VIRU Putative coat protein OS=Heterobasidion partitivirus 2 OX=872291 PE=4 SV=1
MM1 pKa = 7.78ADD3 pKa = 3.44KK4 pKa = 10.76VDD6 pKa = 3.74PPRR9 pKa = 11.84DD10 pKa = 3.7VVAEE14 pKa = 4.35TPPVPQPASSGRR26 pKa = 11.84TVVKK30 pKa = 10.13QRR32 pKa = 11.84HH33 pKa = 5.33PAQGAPTISLAKK45 pKa = 10.65VLDD48 pKa = 4.11LEE50 pKa = 4.6SDD52 pKa = 3.68VHH54 pKa = 7.12KK55 pKa = 11.18GKK57 pKa = 8.74GTYY60 pKa = 9.81HH61 pKa = 6.43SGSYY65 pKa = 10.27VPDD68 pKa = 2.96VRR70 pKa = 11.84FFVLSLCTYY79 pKa = 7.87YY80 pKa = 11.16GRR82 pKa = 11.84VFSATDD88 pKa = 3.38YY89 pKa = 9.55EE90 pKa = 4.42ASQFVTPPSLIAYY103 pKa = 9.48SLFCLYY109 pKa = 10.64FLLFYY114 pKa = 10.85RR115 pKa = 11.84DD116 pKa = 3.7MHH118 pKa = 6.79SKK120 pKa = 10.56SPSQYY125 pKa = 10.72ARR127 pKa = 11.84TFTDD131 pKa = 4.18FGRR134 pKa = 11.84YY135 pKa = 7.94NQLLKK140 pKa = 11.3LMDD143 pKa = 3.74EE144 pKa = 4.64VYY146 pKa = 10.51IPDD149 pKa = 4.68EE150 pKa = 4.01IFEE153 pKa = 4.37LLKK156 pKa = 10.99VFSVWSPEE164 pKa = 4.42LIPNLQFVPSLGSTLPLYY182 pKa = 9.69DD183 pKa = 3.97IPYY186 pKa = 9.62LMHH189 pKa = 6.92PAIFLHH195 pKa = 5.87GHH197 pKa = 4.6NTLFNRR203 pKa = 11.84ADD205 pKa = 3.52AVGHH209 pKa = 4.98YY210 pKa = 10.78ARR212 pKa = 11.84FLRR215 pKa = 11.84TEE217 pKa = 4.16LFSINSASLGNAARR231 pKa = 11.84IMTIGNLIGCAYY243 pKa = 8.88TPNATQGSARR253 pKa = 11.84LIQNWLSNAILPFVDD268 pKa = 3.97PATHH272 pKa = 6.05RR273 pKa = 11.84QHH275 pKa = 6.99LRR277 pKa = 11.84RR278 pKa = 11.84TGLAKK283 pKa = 10.37YY284 pKa = 10.4DD285 pKa = 3.65LGLPTFTSANWNPYY299 pKa = 8.67TFLFSSNVSSSFEE312 pKa = 4.05SFIDD316 pKa = 3.78CAHH319 pKa = 6.65ACSEE323 pKa = 4.34FSRR326 pKa = 11.84DD327 pKa = 3.25QLKK330 pKa = 10.63ASRR333 pKa = 11.84HH334 pKa = 4.43MSDD337 pKa = 4.83LYY339 pKa = 11.12SQVSPAPGSYY349 pKa = 9.65MIQSYY354 pKa = 7.35STPTWHH360 pKa = 6.59ATSLPSASFPDD371 pKa = 4.22GEE373 pKa = 4.3ITRR376 pKa = 11.84GDD378 pKa = 4.2FDD380 pKa = 4.4TLHH383 pKa = 5.13TTYY386 pKa = 10.14TFFGIAPAAATANSNNLHH404 pKa = 5.61MPEE407 pKa = 4.07VRR409 pKa = 11.84QNPATATSTLIGFVPNAYY427 pKa = 10.29LIEE430 pKa = 4.08QHH432 pKa = 7.02ADD434 pKa = 3.09PDD436 pKa = 4.05VAVPQQLSLIDD447 pKa = 3.9RR448 pKa = 11.84NEE450 pKa = 4.2MKK452 pKa = 9.36HH453 pKa = 5.01TRR455 pKa = 11.84PNILVFAPGDD465 pKa = 4.01STVSAAQWPILTGIVVHH482 pKa = 6.53NGNIDD487 pKa = 3.85SNVIKK492 pKa = 10.59APNPDD497 pKa = 3.91DD498 pKa = 5.43DD499 pKa = 4.9LPQIRR504 pKa = 11.84SRR506 pKa = 11.84YY507 pKa = 8.11VDD509 pKa = 3.25GFVSLRR515 pKa = 11.84VIRR518 pKa = 11.84PRR520 pKa = 11.84FANRR524 pKa = 11.84PTWLIARR531 pKa = 11.84QALEE535 pKa = 3.93RR536 pKa = 11.84VSYY539 pKa = 9.31STINFLWRR547 pKa = 11.84SDD549 pKa = 4.15LIAVPRR555 pKa = 11.84FDD557 pKa = 4.28AAHH560 pKa = 6.42NAPAAGGITGLLWPFTFFRR579 pKa = 11.84RR580 pKa = 11.84VTQMIFGTNVAVSDD594 pKa = 4.33SPTDD598 pKa = 3.51PTVASHH604 pKa = 6.12KK605 pKa = 10.09QVGDD609 pKa = 2.88AWSSFRR615 pKa = 11.84LTSDD619 pKa = 3.22SNAVPSVEE627 pKa = 3.65NTYY630 pKa = 10.86FGINHH635 pKa = 6.5GEE637 pKa = 3.93IIFGRR642 pKa = 11.84NSTLNKK648 pKa = 9.8FSHH651 pKa = 6.78PSDD654 pKa = 3.42LLRR657 pKa = 11.84RR658 pKa = 11.84NN659 pKa = 3.45
MM1 pKa = 7.78ADD3 pKa = 3.44KK4 pKa = 10.76VDD6 pKa = 3.74PPRR9 pKa = 11.84DD10 pKa = 3.7VVAEE14 pKa = 4.35TPPVPQPASSGRR26 pKa = 11.84TVVKK30 pKa = 10.13QRR32 pKa = 11.84HH33 pKa = 5.33PAQGAPTISLAKK45 pKa = 10.65VLDD48 pKa = 4.11LEE50 pKa = 4.6SDD52 pKa = 3.68VHH54 pKa = 7.12KK55 pKa = 11.18GKK57 pKa = 8.74GTYY60 pKa = 9.81HH61 pKa = 6.43SGSYY65 pKa = 10.27VPDD68 pKa = 2.96VRR70 pKa = 11.84FFVLSLCTYY79 pKa = 7.87YY80 pKa = 11.16GRR82 pKa = 11.84VFSATDD88 pKa = 3.38YY89 pKa = 9.55EE90 pKa = 4.42ASQFVTPPSLIAYY103 pKa = 9.48SLFCLYY109 pKa = 10.64FLLFYY114 pKa = 10.85RR115 pKa = 11.84DD116 pKa = 3.7MHH118 pKa = 6.79SKK120 pKa = 10.56SPSQYY125 pKa = 10.72ARR127 pKa = 11.84TFTDD131 pKa = 4.18FGRR134 pKa = 11.84YY135 pKa = 7.94NQLLKK140 pKa = 11.3LMDD143 pKa = 3.74EE144 pKa = 4.64VYY146 pKa = 10.51IPDD149 pKa = 4.68EE150 pKa = 4.01IFEE153 pKa = 4.37LLKK156 pKa = 10.99VFSVWSPEE164 pKa = 4.42LIPNLQFVPSLGSTLPLYY182 pKa = 9.69DD183 pKa = 3.97IPYY186 pKa = 9.62LMHH189 pKa = 6.92PAIFLHH195 pKa = 5.87GHH197 pKa = 4.6NTLFNRR203 pKa = 11.84ADD205 pKa = 3.52AVGHH209 pKa = 4.98YY210 pKa = 10.78ARR212 pKa = 11.84FLRR215 pKa = 11.84TEE217 pKa = 4.16LFSINSASLGNAARR231 pKa = 11.84IMTIGNLIGCAYY243 pKa = 8.88TPNATQGSARR253 pKa = 11.84LIQNWLSNAILPFVDD268 pKa = 3.97PATHH272 pKa = 6.05RR273 pKa = 11.84QHH275 pKa = 6.99LRR277 pKa = 11.84RR278 pKa = 11.84TGLAKK283 pKa = 10.37YY284 pKa = 10.4DD285 pKa = 3.65LGLPTFTSANWNPYY299 pKa = 8.67TFLFSSNVSSSFEE312 pKa = 4.05SFIDD316 pKa = 3.78CAHH319 pKa = 6.65ACSEE323 pKa = 4.34FSRR326 pKa = 11.84DD327 pKa = 3.25QLKK330 pKa = 10.63ASRR333 pKa = 11.84HH334 pKa = 4.43MSDD337 pKa = 4.83LYY339 pKa = 11.12SQVSPAPGSYY349 pKa = 9.65MIQSYY354 pKa = 7.35STPTWHH360 pKa = 6.59ATSLPSASFPDD371 pKa = 4.22GEE373 pKa = 4.3ITRR376 pKa = 11.84GDD378 pKa = 4.2FDD380 pKa = 4.4TLHH383 pKa = 5.13TTYY386 pKa = 10.14TFFGIAPAAATANSNNLHH404 pKa = 5.61MPEE407 pKa = 4.07VRR409 pKa = 11.84QNPATATSTLIGFVPNAYY427 pKa = 10.29LIEE430 pKa = 4.08QHH432 pKa = 7.02ADD434 pKa = 3.09PDD436 pKa = 4.05VAVPQQLSLIDD447 pKa = 3.9RR448 pKa = 11.84NEE450 pKa = 4.2MKK452 pKa = 9.36HH453 pKa = 5.01TRR455 pKa = 11.84PNILVFAPGDD465 pKa = 4.01STVSAAQWPILTGIVVHH482 pKa = 6.53NGNIDD487 pKa = 3.85SNVIKK492 pKa = 10.59APNPDD497 pKa = 3.91DD498 pKa = 5.43DD499 pKa = 4.9LPQIRR504 pKa = 11.84SRR506 pKa = 11.84YY507 pKa = 8.11VDD509 pKa = 3.25GFVSLRR515 pKa = 11.84VIRR518 pKa = 11.84PRR520 pKa = 11.84FANRR524 pKa = 11.84PTWLIARR531 pKa = 11.84QALEE535 pKa = 3.93RR536 pKa = 11.84VSYY539 pKa = 9.31STINFLWRR547 pKa = 11.84SDD549 pKa = 4.15LIAVPRR555 pKa = 11.84FDD557 pKa = 4.28AAHH560 pKa = 6.42NAPAAGGITGLLWPFTFFRR579 pKa = 11.84RR580 pKa = 11.84VTQMIFGTNVAVSDD594 pKa = 4.33SPTDD598 pKa = 3.51PTVASHH604 pKa = 6.12KK605 pKa = 10.09QVGDD609 pKa = 2.88AWSSFRR615 pKa = 11.84LTSDD619 pKa = 3.22SNAVPSVEE627 pKa = 3.65NTYY630 pKa = 10.86FGINHH635 pKa = 6.5GEE637 pKa = 3.93IIFGRR642 pKa = 11.84NSTLNKK648 pKa = 9.8FSHH651 pKa = 6.78PSDD654 pKa = 3.42LLRR657 pKa = 11.84RR658 pKa = 11.84NN659 pKa = 3.45
Molecular weight: 73.23 kDa
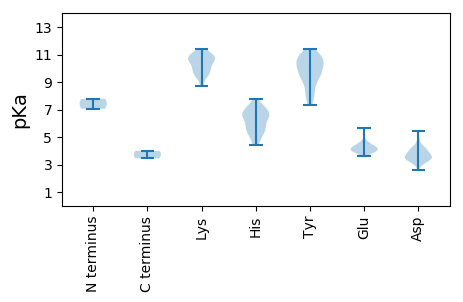
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1381 |
659 |
722 |
690.5 |
79.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.024 ± 1.111 | 1.159 ± 0.273 |
6.155 ± 0.369 | 3.186 ± 0.415 |
6.951 ± 0.499 | 4.345 ± 0.453 |
3.838 ± 0.341 | 5.865 ± 0.379 |
3.476 ± 0.924 | 9.486 ± 0.468 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.245 ± 0.497 | 4.779 ± 0.26 |
6.517 ± 0.732 | 3.765 ± 0.292 |
5.576 ± 0.13 | 7.893 ± 1.243 |
5.793 ± 0.812 | 5.576 ± 0.545 |
1.593 ± 0.155 | 4.779 ± 0.674 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
