
Thioalbus denitrificans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Thioalbus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
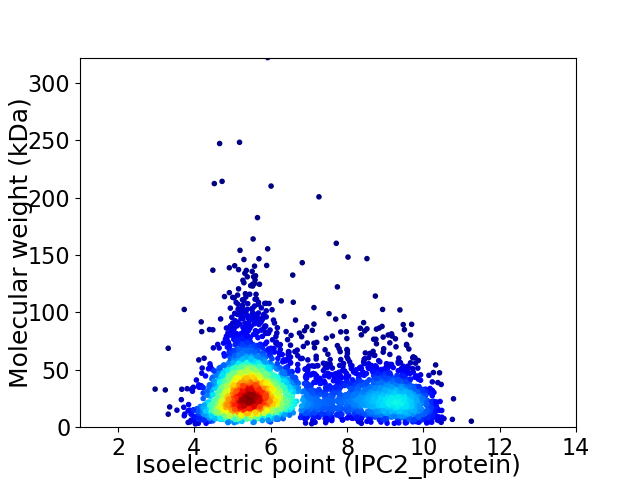
Virtual 2D-PAGE plot for 3827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369C7S2|A0A369C7S2_9GAMM Ankyrin repeat protein OS=Thioalbus denitrificans OX=547122 GN=DFQ59_10635 PE=4 SV=1
MM1 pKa = 6.95EE2 pKa = 4.9TSTPSKK8 pKa = 10.15NSRR11 pKa = 11.84AIWLASICLLVGVLGFGAVPASAASGIFTRR41 pKa = 11.84IFEE44 pKa = 4.35MEE46 pKa = 3.94GNAIDD51 pKa = 4.54EE52 pKa = 4.67NDD54 pKa = 3.16ASLYY58 pKa = 10.81KK59 pKa = 10.58EE60 pKa = 4.31GEE62 pKa = 3.73DD63 pKa = 3.24WQRR66 pKa = 11.84TNNDD70 pKa = 2.06IWVDD74 pKa = 3.2LWNTYY79 pKa = 10.06LPSATKK85 pKa = 8.66PTGWDD90 pKa = 2.89FDD92 pKa = 3.87VPYY95 pKa = 10.15SGKK98 pKa = 9.54AAVRR102 pKa = 11.84VFVPDD107 pKa = 3.59YY108 pKa = 9.91PGLPGFVGTIFDD120 pKa = 4.84DD121 pKa = 4.59NIFGAGCKK129 pKa = 9.84DD130 pKa = 3.69DD131 pKa = 6.19LDD133 pKa = 5.73LNTCKK138 pKa = 10.49SEE140 pKa = 4.22TGNSPDD146 pKa = 3.88KK147 pKa = 11.03NQWTNGYY154 pKa = 9.55AAFYY158 pKa = 10.19NIEE161 pKa = 4.05GLAAPVPVDD170 pKa = 3.44GLPPGTLYY178 pKa = 10.46HH179 pKa = 6.26QNGDD183 pKa = 3.66LVIVMGGDD191 pKa = 3.27VHH193 pKa = 8.54DD194 pKa = 5.38GDD196 pKa = 5.13GSSQVGGWLFRR207 pKa = 11.84QQVPAEE213 pKa = 4.33LIVPGEE219 pKa = 4.36DD220 pKa = 3.51FPFSRR225 pKa = 11.84SIGDD229 pKa = 3.38ILVTVDD235 pKa = 2.81WSEE238 pKa = 4.14GGRR241 pKa = 11.84TATVRR246 pKa = 11.84AYY248 pKa = 10.37RR249 pKa = 11.84WIGGDD254 pKa = 3.28TTEE257 pKa = 4.95CPTPGVVLGTDD268 pKa = 3.76DD269 pKa = 4.18NLCKK273 pKa = 10.27IFEE276 pKa = 4.66GPNAVCTPEE285 pKa = 3.91YY286 pKa = 10.8NGLACAIANYY296 pKa = 10.35APTEE300 pKa = 3.94EE301 pKa = 4.33APEE304 pKa = 4.49GVGEE308 pKa = 4.67TPAPWPYY315 pKa = 9.53IAKK318 pKa = 9.85GDD320 pKa = 3.98RR321 pKa = 11.84SVDD324 pKa = 3.38PANGIFTEE332 pKa = 4.34NFPEE336 pKa = 4.29TTLLEE341 pKa = 5.02GILNLSAILRR351 pKa = 11.84LNDD354 pKa = 3.21EE355 pKa = 4.61TVGCFSDD362 pKa = 5.35FMWEE366 pKa = 4.22TRR368 pKa = 11.84SSGAPGEE375 pKa = 4.36TYY377 pKa = 9.91NAKK380 pKa = 10.59LKK382 pKa = 10.7DD383 pKa = 3.84YY384 pKa = 11.0LLGDD388 pKa = 3.92TEE390 pKa = 4.64LCGIDD395 pKa = 3.48VDD397 pKa = 4.47KK398 pKa = 11.7SGGTLSKK405 pKa = 11.16VGDD408 pKa = 3.69LVSYY412 pKa = 10.71NILVEE417 pKa = 3.83NTGIVRR423 pKa = 11.84LYY425 pKa = 9.75KK426 pKa = 10.5QSITDD431 pKa = 3.62TLLGDD436 pKa = 4.04LTSTNGNCPGGDD448 pKa = 3.54EE449 pKa = 5.18DD450 pKa = 5.59NLTEE454 pKa = 4.65GEE456 pKa = 4.42SCVEE460 pKa = 4.24SNACGAYY467 pKa = 10.31LEE469 pKa = 4.87PDD471 pKa = 3.74GTCAITAWRR480 pKa = 11.84RR481 pKa = 11.84VATGDD486 pKa = 4.28PDD488 pKa = 4.13PLPNTVTVVYY498 pKa = 10.59DD499 pKa = 3.98SDD501 pKa = 4.15PGLQGDD507 pKa = 4.67EE508 pKa = 4.19VTSSNDD514 pKa = 2.82HH515 pKa = 6.09SVNLFQPAVEE525 pKa = 4.28VVKK528 pKa = 11.17GGDD531 pKa = 3.64TLSKK535 pKa = 10.98AGDD538 pKa = 3.59DD539 pKa = 3.58VTYY542 pKa = 11.16SFTINNLSSDD552 pKa = 4.07DD553 pKa = 4.21APNLLIDD560 pKa = 4.29DD561 pKa = 4.35TSGVTDD567 pKa = 3.62TVLGNLTATAKK578 pKa = 11.02ANGCTSLAYY587 pKa = 10.16GGSCSFNVVYY597 pKa = 9.18TVQSGDD603 pKa = 4.24PDD605 pKa = 4.13PLDD608 pKa = 3.44NTVSVLYY615 pKa = 10.71HH616 pKa = 6.9PDD618 pKa = 3.31GFPNNITDD626 pKa = 3.87EE627 pKa = 4.72DD628 pKa = 4.18SHH630 pKa = 8.01SVNLFQPSVDD640 pKa = 3.49VTKK643 pKa = 10.58TGDD646 pKa = 3.51TLSKK650 pKa = 11.03VGDD653 pKa = 3.77PVNYY657 pKa = 9.55TITVTNDD664 pKa = 2.78GSSDD668 pKa = 3.68NPGLEE673 pKa = 4.12NGTITDD679 pKa = 3.87TLLGNLLDD687 pKa = 4.24GSNPYY692 pKa = 8.2VTSSDD697 pKa = 3.59CTGTLGPSGSCTIQATRR714 pKa = 11.84TVQAGDD720 pKa = 4.24PDD722 pKa = 4.55PLPNTVTVHH731 pKa = 6.46FDD733 pKa = 3.25PTGSFDD739 pKa = 4.94NDD741 pKa = 2.95ITDD744 pKa = 3.76SDD746 pKa = 3.9GHH748 pKa = 5.8SVNLFQPGVDD758 pKa = 3.51VTKK761 pKa = 10.59TGDD764 pKa = 3.51TLSKK768 pKa = 10.94VGDD771 pKa = 3.69PASYY775 pKa = 9.97TITVNNTGSGDD786 pKa = 3.77SPNLVNGTISDD797 pKa = 3.99TLLGNLLDD805 pKa = 4.24GSNPYY810 pKa = 8.27VTSSTCSATLATGASCTIQATRR832 pKa = 11.84TVQAGDD838 pKa = 4.24PDD840 pKa = 4.55PLPNTVTVHH849 pKa = 5.52YY850 pKa = 10.62NPDD853 pKa = 3.4GFPNDD858 pKa = 3.34ISDD861 pKa = 4.19SDD863 pKa = 3.75GHH865 pKa = 5.89SVNLFQPGVDD875 pKa = 3.51VTKK878 pKa = 10.59TGDD881 pKa = 3.51TLSKK885 pKa = 10.94VGDD888 pKa = 3.69PASYY892 pKa = 9.97TITVNNTGSGDD903 pKa = 3.77SPNLVNGTISDD914 pKa = 3.99TLLGNLLDD922 pKa = 4.24GSNPYY927 pKa = 8.27VTSSTCSAALATGASCTIQATRR949 pKa = 11.84TVQAGDD955 pKa = 4.24PDD957 pKa = 4.55PLPNTVTVHH966 pKa = 5.52YY967 pKa = 10.62NPDD970 pKa = 3.4GFPNDD975 pKa = 3.82ISDD978 pKa = 3.8STT980 pKa = 3.85
MM1 pKa = 6.95EE2 pKa = 4.9TSTPSKK8 pKa = 10.15NSRR11 pKa = 11.84AIWLASICLLVGVLGFGAVPASAASGIFTRR41 pKa = 11.84IFEE44 pKa = 4.35MEE46 pKa = 3.94GNAIDD51 pKa = 4.54EE52 pKa = 4.67NDD54 pKa = 3.16ASLYY58 pKa = 10.81KK59 pKa = 10.58EE60 pKa = 4.31GEE62 pKa = 3.73DD63 pKa = 3.24WQRR66 pKa = 11.84TNNDD70 pKa = 2.06IWVDD74 pKa = 3.2LWNTYY79 pKa = 10.06LPSATKK85 pKa = 8.66PTGWDD90 pKa = 2.89FDD92 pKa = 3.87VPYY95 pKa = 10.15SGKK98 pKa = 9.54AAVRR102 pKa = 11.84VFVPDD107 pKa = 3.59YY108 pKa = 9.91PGLPGFVGTIFDD120 pKa = 4.84DD121 pKa = 4.59NIFGAGCKK129 pKa = 9.84DD130 pKa = 3.69DD131 pKa = 6.19LDD133 pKa = 5.73LNTCKK138 pKa = 10.49SEE140 pKa = 4.22TGNSPDD146 pKa = 3.88KK147 pKa = 11.03NQWTNGYY154 pKa = 9.55AAFYY158 pKa = 10.19NIEE161 pKa = 4.05GLAAPVPVDD170 pKa = 3.44GLPPGTLYY178 pKa = 10.46HH179 pKa = 6.26QNGDD183 pKa = 3.66LVIVMGGDD191 pKa = 3.27VHH193 pKa = 8.54DD194 pKa = 5.38GDD196 pKa = 5.13GSSQVGGWLFRR207 pKa = 11.84QQVPAEE213 pKa = 4.33LIVPGEE219 pKa = 4.36DD220 pKa = 3.51FPFSRR225 pKa = 11.84SIGDD229 pKa = 3.38ILVTVDD235 pKa = 2.81WSEE238 pKa = 4.14GGRR241 pKa = 11.84TATVRR246 pKa = 11.84AYY248 pKa = 10.37RR249 pKa = 11.84WIGGDD254 pKa = 3.28TTEE257 pKa = 4.95CPTPGVVLGTDD268 pKa = 3.76DD269 pKa = 4.18NLCKK273 pKa = 10.27IFEE276 pKa = 4.66GPNAVCTPEE285 pKa = 3.91YY286 pKa = 10.8NGLACAIANYY296 pKa = 10.35APTEE300 pKa = 3.94EE301 pKa = 4.33APEE304 pKa = 4.49GVGEE308 pKa = 4.67TPAPWPYY315 pKa = 9.53IAKK318 pKa = 9.85GDD320 pKa = 3.98RR321 pKa = 11.84SVDD324 pKa = 3.38PANGIFTEE332 pKa = 4.34NFPEE336 pKa = 4.29TTLLEE341 pKa = 5.02GILNLSAILRR351 pKa = 11.84LNDD354 pKa = 3.21EE355 pKa = 4.61TVGCFSDD362 pKa = 5.35FMWEE366 pKa = 4.22TRR368 pKa = 11.84SSGAPGEE375 pKa = 4.36TYY377 pKa = 9.91NAKK380 pKa = 10.59LKK382 pKa = 10.7DD383 pKa = 3.84YY384 pKa = 11.0LLGDD388 pKa = 3.92TEE390 pKa = 4.64LCGIDD395 pKa = 3.48VDD397 pKa = 4.47KK398 pKa = 11.7SGGTLSKK405 pKa = 11.16VGDD408 pKa = 3.69LVSYY412 pKa = 10.71NILVEE417 pKa = 3.83NTGIVRR423 pKa = 11.84LYY425 pKa = 9.75KK426 pKa = 10.5QSITDD431 pKa = 3.62TLLGDD436 pKa = 4.04LTSTNGNCPGGDD448 pKa = 3.54EE449 pKa = 5.18DD450 pKa = 5.59NLTEE454 pKa = 4.65GEE456 pKa = 4.42SCVEE460 pKa = 4.24SNACGAYY467 pKa = 10.31LEE469 pKa = 4.87PDD471 pKa = 3.74GTCAITAWRR480 pKa = 11.84RR481 pKa = 11.84VATGDD486 pKa = 4.28PDD488 pKa = 4.13PLPNTVTVVYY498 pKa = 10.59DD499 pKa = 3.98SDD501 pKa = 4.15PGLQGDD507 pKa = 4.67EE508 pKa = 4.19VTSSNDD514 pKa = 2.82HH515 pKa = 6.09SVNLFQPAVEE525 pKa = 4.28VVKK528 pKa = 11.17GGDD531 pKa = 3.64TLSKK535 pKa = 10.98AGDD538 pKa = 3.59DD539 pKa = 3.58VTYY542 pKa = 11.16SFTINNLSSDD552 pKa = 4.07DD553 pKa = 4.21APNLLIDD560 pKa = 4.29DD561 pKa = 4.35TSGVTDD567 pKa = 3.62TVLGNLTATAKK578 pKa = 11.02ANGCTSLAYY587 pKa = 10.16GGSCSFNVVYY597 pKa = 9.18TVQSGDD603 pKa = 4.24PDD605 pKa = 4.13PLDD608 pKa = 3.44NTVSVLYY615 pKa = 10.71HH616 pKa = 6.9PDD618 pKa = 3.31GFPNNITDD626 pKa = 3.87EE627 pKa = 4.72DD628 pKa = 4.18SHH630 pKa = 8.01SVNLFQPSVDD640 pKa = 3.49VTKK643 pKa = 10.58TGDD646 pKa = 3.51TLSKK650 pKa = 11.03VGDD653 pKa = 3.77PVNYY657 pKa = 9.55TITVTNDD664 pKa = 2.78GSSDD668 pKa = 3.68NPGLEE673 pKa = 4.12NGTITDD679 pKa = 3.87TLLGNLLDD687 pKa = 4.24GSNPYY692 pKa = 8.2VTSSDD697 pKa = 3.59CTGTLGPSGSCTIQATRR714 pKa = 11.84TVQAGDD720 pKa = 4.24PDD722 pKa = 4.55PLPNTVTVHH731 pKa = 6.46FDD733 pKa = 3.25PTGSFDD739 pKa = 4.94NDD741 pKa = 2.95ITDD744 pKa = 3.76SDD746 pKa = 3.9GHH748 pKa = 5.8SVNLFQPGVDD758 pKa = 3.51VTKK761 pKa = 10.59TGDD764 pKa = 3.51TLSKK768 pKa = 10.94VGDD771 pKa = 3.69PASYY775 pKa = 9.97TITVNNTGSGDD786 pKa = 3.77SPNLVNGTISDD797 pKa = 3.99TLLGNLLDD805 pKa = 4.24GSNPYY810 pKa = 8.27VTSSTCSATLATGASCTIQATRR832 pKa = 11.84TVQAGDD838 pKa = 4.24PDD840 pKa = 4.55PLPNTVTVHH849 pKa = 5.52YY850 pKa = 10.62NPDD853 pKa = 3.4GFPNDD858 pKa = 3.34ISDD861 pKa = 4.19SDD863 pKa = 3.75GHH865 pKa = 5.89SVNLFQPGVDD875 pKa = 3.51VTKK878 pKa = 10.59TGDD881 pKa = 3.51TLSKK885 pKa = 10.94VGDD888 pKa = 3.69PASYY892 pKa = 9.97TITVNNTGSGDD903 pKa = 3.77SPNLVNGTISDD914 pKa = 3.99TLLGNLLDD922 pKa = 4.24GSNPYY927 pKa = 8.27VTSSTCSAALATGASCTIQATRR949 pKa = 11.84TVQAGDD955 pKa = 4.24PDD957 pKa = 4.55PLPNTVTVHH966 pKa = 5.52YY967 pKa = 10.62NPDD970 pKa = 3.4GFPNDD975 pKa = 3.82ISDD978 pKa = 3.8STT980 pKa = 3.85
Molecular weight: 102.59 kDa
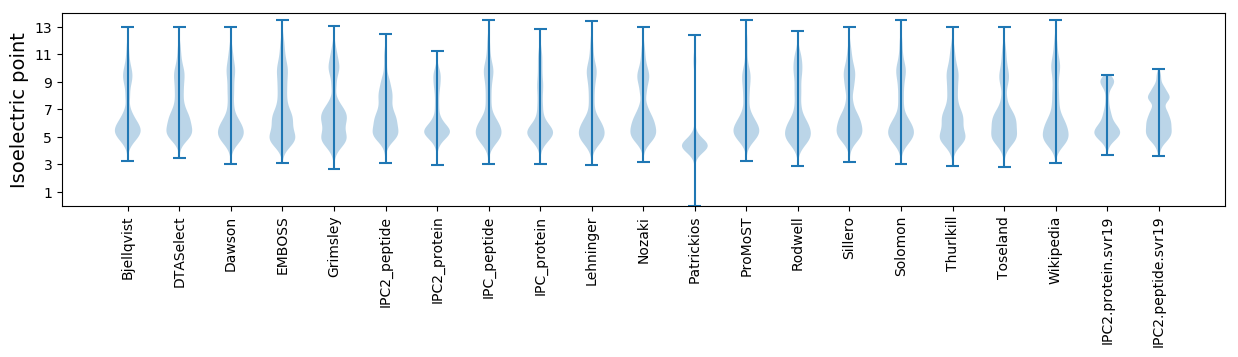
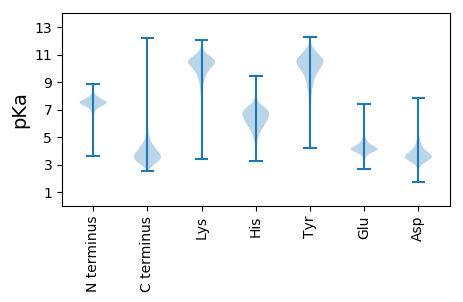
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369CH38|A0A369CH38_9GAMM DinB family protein OS=Thioalbus denitrificans OX=547122 GN=DFQ59_102144 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84QIIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33NGRR28 pKa = 11.84QIIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1199143 |
29 |
2919 |
313.3 |
34.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.957 ± 0.055 | 0.98 ± 0.015 |
5.306 ± 0.031 | 6.628 ± 0.04 |
3.347 ± 0.022 | 9.124 ± 0.04 |
2.313 ± 0.021 | 4.147 ± 0.032 |
2.324 ± 0.034 | 11.576 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.194 ± 0.021 | 2.327 ± 0.021 |
5.507 ± 0.035 | 3.078 ± 0.027 |
8.375 ± 0.044 | 4.544 ± 0.025 |
4.83 ± 0.023 | 7.565 ± 0.032 |
1.42 ± 0.018 | 2.458 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
