
Spathaspora passalidarum (strain NRRL Y-27907 / 11-Y1)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Spathaspora; Spathaspora passalidarum
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
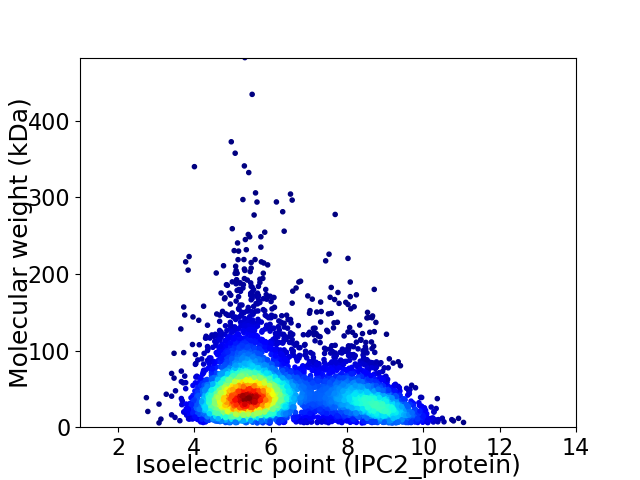
Virtual 2D-PAGE plot for 5973 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G3AGF1|G3AGF1_SPAPN Acyl carrier protein OS=Spathaspora passalidarum (strain NRRL Y-27907 / 11-Y1) OX=619300 GN=SPAPADRAFT_58508 PE=3 SV=1
MM1 pKa = 7.24YY2 pKa = 10.63LLLILLQLLTFAATQEE18 pKa = 3.84ISGVFTSFNSLTWLNSNPNTFQTPASPAWIAEE50 pKa = 4.28LSWKK54 pKa = 10.0IDD56 pKa = 3.42GSVVSAGDD64 pKa = 3.72TFSLTMPCVFRR75 pKa = 11.84FYY77 pKa = 10.78TDD79 pKa = 3.05QDD81 pKa = 4.03SVNLRR86 pKa = 11.84VGSTNYY92 pKa = 8.08ATCDD96 pKa = 4.44FITGGISSTDD106 pKa = 3.3SEE108 pKa = 5.06LNCVLSDD115 pKa = 3.69EE116 pKa = 4.5VEE118 pKa = 4.04EE119 pKa = 4.98SILAVGKK126 pKa = 8.69LTFPIVFNIGRR137 pKa = 11.84STEE140 pKa = 4.78DD141 pKa = 2.78VDD143 pKa = 5.68LYY145 pKa = 11.35CSTFYY150 pKa = 10.65QGGLNTISFMDD161 pKa = 4.03GDD163 pKa = 4.08NTLSIDD169 pKa = 4.33VEE171 pKa = 4.48FQDD174 pKa = 4.69GTPSGDD180 pKa = 3.7PNGPMYY186 pKa = 10.25GARR189 pKa = 11.84MLPSSNRR196 pKa = 11.84WQNYY200 pKa = 6.84VLGGLCPGGLSTATIGITINDD221 pKa = 4.16DD222 pKa = 3.16VSTIDD227 pKa = 5.04CEE229 pKa = 4.27TANIRR234 pKa = 11.84LTNAVNDD241 pKa = 3.65WFYY244 pKa = 11.03PEE246 pKa = 4.13SFKK249 pKa = 10.98TFTTASKK256 pKa = 10.56LCRR259 pKa = 11.84SNSFMITYY267 pKa = 9.33WGVPDD272 pKa = 4.67GYY274 pKa = 10.65RR275 pKa = 11.84VFIEE279 pKa = 4.42AVINTDD285 pKa = 3.26GNPPRR290 pKa = 11.84VQYY293 pKa = 8.82TNKK296 pKa = 9.55YY297 pKa = 9.15LCPTTIDD304 pKa = 3.11NGVWVNWLPYY314 pKa = 10.61DD315 pKa = 3.82SVEE318 pKa = 3.95ADD320 pKa = 3.56GPGKK324 pKa = 10.38EE325 pKa = 4.35IITVTEE331 pKa = 3.92TWTGSDD337 pKa = 3.43TEE339 pKa = 4.77TITKK343 pKa = 10.43SFDD346 pKa = 3.19EE347 pKa = 5.6DD348 pKa = 3.5EE349 pKa = 5.14DD350 pKa = 3.89LTITIAVNIPIPTITLTEE368 pKa = 4.33TYY370 pKa = 10.86VGITTSYY377 pKa = 6.95TTYY380 pKa = 9.58TVEE383 pKa = 4.3PGEE386 pKa = 4.24TASVVVYY393 pKa = 10.51DD394 pKa = 4.68PVHH397 pKa = 4.66TTITEE402 pKa = 4.11EE403 pKa = 4.33SCWEE407 pKa = 3.89SDD409 pKa = 3.28EE410 pKa = 5.94SITTTIIDD418 pKa = 4.58PDD420 pKa = 3.79WASDD424 pKa = 3.64TILIVTPCEE433 pKa = 4.12VTSTEE438 pKa = 4.09QEE440 pKa = 3.92EE441 pKa = 4.93SSTKK445 pKa = 10.37VEE447 pKa = 5.54DD448 pKa = 3.74IPSDD452 pKa = 3.59DD453 pKa = 4.05TLDD456 pKa = 4.25DD457 pKa = 4.48DD458 pKa = 5.46SYY460 pKa = 11.7NPPEE464 pKa = 4.17IADD467 pKa = 3.62NASSEE472 pKa = 4.5EE473 pKa = 4.07ITSGEE478 pKa = 4.18VQSTEE483 pKa = 3.75ILSSEE488 pKa = 4.29EE489 pKa = 4.03VQSTEE494 pKa = 3.73ILSSEE499 pKa = 4.29EE500 pKa = 4.03VQSTEE505 pKa = 3.8ILSSEE510 pKa = 4.11EE511 pKa = 4.38SFSQDD516 pKa = 2.55STTVPYY522 pKa = 10.89GSTKK526 pKa = 7.91EE527 pKa = 4.31TEE529 pKa = 3.7ISTEE533 pKa = 3.69IVLEE537 pKa = 4.3EE538 pKa = 4.23TFSTEE543 pKa = 4.15DD544 pKa = 3.25TTPEE548 pKa = 3.74NNTWDD553 pKa = 4.1GLTDD557 pKa = 5.09DD558 pKa = 6.06DD559 pKa = 4.79SQQPSSNTKK568 pKa = 8.79NDD570 pKa = 3.39SNEE573 pKa = 4.28EE574 pKa = 4.08YY575 pKa = 10.4TSDD578 pKa = 4.12QIDD581 pKa = 3.41ISSSGSEE588 pKa = 3.86TSAKK592 pKa = 8.72QTTTEE597 pKa = 4.53TIPDD601 pKa = 3.42NHH603 pKa = 6.29TTDD606 pKa = 4.0IVQEE610 pKa = 4.21DD611 pKa = 4.52PGTEE615 pKa = 3.64ILSVEE620 pKa = 4.19NTTVRR625 pKa = 11.84VPDD628 pKa = 4.85DD629 pKa = 3.67NTTDD633 pKa = 3.27ISPSDD638 pKa = 3.85VICNSEE644 pKa = 4.1NEE646 pKa = 4.33ASSEE650 pKa = 4.01PNEE653 pKa = 3.71TRR655 pKa = 11.84TYY657 pKa = 10.59VSIVPTTKK665 pKa = 10.63SKK667 pKa = 11.36DD668 pKa = 3.5STITSAFSGYY678 pKa = 9.74EE679 pKa = 3.27QDD681 pKa = 4.15TITKK685 pKa = 9.5VITYY689 pKa = 7.28TTTICEE695 pKa = 4.51SDD697 pKa = 3.46TVTQVITYY705 pKa = 6.74TTIITNHH712 pKa = 6.63DD713 pKa = 4.59SIAKK717 pKa = 7.96TATEE721 pKa = 4.23MTTEE725 pKa = 3.99ATSFAVTTVGEE736 pKa = 4.34TRR738 pKa = 11.84TIIYY742 pKa = 9.54LQDD745 pKa = 3.32SMGTTNDD752 pKa = 3.15TGNNSSNTKK761 pKa = 9.86SDD763 pKa = 2.98SRR765 pKa = 11.84TASNNTGNNSSNTKK779 pKa = 9.92SYY781 pKa = 11.19SGTASNEE788 pKa = 4.14EE789 pKa = 3.93IQTTANSEE797 pKa = 4.18NNSEE801 pKa = 4.22HH802 pKa = 6.07TKK804 pKa = 10.56SEE806 pKa = 4.14SGQSDD811 pKa = 2.61ISEE814 pKa = 4.49YY815 pKa = 10.86YY816 pKa = 10.52SGGTEE821 pKa = 3.9SDD823 pKa = 3.19SGKK826 pKa = 10.47GNKK829 pKa = 9.52SVVPEE834 pKa = 4.16EE835 pKa = 4.09NQGTNSRR842 pKa = 11.84NNNSSTNSNNRR853 pKa = 11.84SDD855 pKa = 4.41DD856 pKa = 3.26QTTRR860 pKa = 11.84GSYY863 pKa = 8.49STSLQLVTSNGIGSTRR879 pKa = 11.84GTSTFSLLFTFVIAILYY896 pKa = 8.94
MM1 pKa = 7.24YY2 pKa = 10.63LLLILLQLLTFAATQEE18 pKa = 3.84ISGVFTSFNSLTWLNSNPNTFQTPASPAWIAEE50 pKa = 4.28LSWKK54 pKa = 10.0IDD56 pKa = 3.42GSVVSAGDD64 pKa = 3.72TFSLTMPCVFRR75 pKa = 11.84FYY77 pKa = 10.78TDD79 pKa = 3.05QDD81 pKa = 4.03SVNLRR86 pKa = 11.84VGSTNYY92 pKa = 8.08ATCDD96 pKa = 4.44FITGGISSTDD106 pKa = 3.3SEE108 pKa = 5.06LNCVLSDD115 pKa = 3.69EE116 pKa = 4.5VEE118 pKa = 4.04EE119 pKa = 4.98SILAVGKK126 pKa = 8.69LTFPIVFNIGRR137 pKa = 11.84STEE140 pKa = 4.78DD141 pKa = 2.78VDD143 pKa = 5.68LYY145 pKa = 11.35CSTFYY150 pKa = 10.65QGGLNTISFMDD161 pKa = 4.03GDD163 pKa = 4.08NTLSIDD169 pKa = 4.33VEE171 pKa = 4.48FQDD174 pKa = 4.69GTPSGDD180 pKa = 3.7PNGPMYY186 pKa = 10.25GARR189 pKa = 11.84MLPSSNRR196 pKa = 11.84WQNYY200 pKa = 6.84VLGGLCPGGLSTATIGITINDD221 pKa = 4.16DD222 pKa = 3.16VSTIDD227 pKa = 5.04CEE229 pKa = 4.27TANIRR234 pKa = 11.84LTNAVNDD241 pKa = 3.65WFYY244 pKa = 11.03PEE246 pKa = 4.13SFKK249 pKa = 10.98TFTTASKK256 pKa = 10.56LCRR259 pKa = 11.84SNSFMITYY267 pKa = 9.33WGVPDD272 pKa = 4.67GYY274 pKa = 10.65RR275 pKa = 11.84VFIEE279 pKa = 4.42AVINTDD285 pKa = 3.26GNPPRR290 pKa = 11.84VQYY293 pKa = 8.82TNKK296 pKa = 9.55YY297 pKa = 9.15LCPTTIDD304 pKa = 3.11NGVWVNWLPYY314 pKa = 10.61DD315 pKa = 3.82SVEE318 pKa = 3.95ADD320 pKa = 3.56GPGKK324 pKa = 10.38EE325 pKa = 4.35IITVTEE331 pKa = 3.92TWTGSDD337 pKa = 3.43TEE339 pKa = 4.77TITKK343 pKa = 10.43SFDD346 pKa = 3.19EE347 pKa = 5.6DD348 pKa = 3.5EE349 pKa = 5.14DD350 pKa = 3.89LTITIAVNIPIPTITLTEE368 pKa = 4.33TYY370 pKa = 10.86VGITTSYY377 pKa = 6.95TTYY380 pKa = 9.58TVEE383 pKa = 4.3PGEE386 pKa = 4.24TASVVVYY393 pKa = 10.51DD394 pKa = 4.68PVHH397 pKa = 4.66TTITEE402 pKa = 4.11EE403 pKa = 4.33SCWEE407 pKa = 3.89SDD409 pKa = 3.28EE410 pKa = 5.94SITTTIIDD418 pKa = 4.58PDD420 pKa = 3.79WASDD424 pKa = 3.64TILIVTPCEE433 pKa = 4.12VTSTEE438 pKa = 4.09QEE440 pKa = 3.92EE441 pKa = 4.93SSTKK445 pKa = 10.37VEE447 pKa = 5.54DD448 pKa = 3.74IPSDD452 pKa = 3.59DD453 pKa = 4.05TLDD456 pKa = 4.25DD457 pKa = 4.48DD458 pKa = 5.46SYY460 pKa = 11.7NPPEE464 pKa = 4.17IADD467 pKa = 3.62NASSEE472 pKa = 4.5EE473 pKa = 4.07ITSGEE478 pKa = 4.18VQSTEE483 pKa = 3.75ILSSEE488 pKa = 4.29EE489 pKa = 4.03VQSTEE494 pKa = 3.73ILSSEE499 pKa = 4.29EE500 pKa = 4.03VQSTEE505 pKa = 3.8ILSSEE510 pKa = 4.11EE511 pKa = 4.38SFSQDD516 pKa = 2.55STTVPYY522 pKa = 10.89GSTKK526 pKa = 7.91EE527 pKa = 4.31TEE529 pKa = 3.7ISTEE533 pKa = 3.69IVLEE537 pKa = 4.3EE538 pKa = 4.23TFSTEE543 pKa = 4.15DD544 pKa = 3.25TTPEE548 pKa = 3.74NNTWDD553 pKa = 4.1GLTDD557 pKa = 5.09DD558 pKa = 6.06DD559 pKa = 4.79SQQPSSNTKK568 pKa = 8.79NDD570 pKa = 3.39SNEE573 pKa = 4.28EE574 pKa = 4.08YY575 pKa = 10.4TSDD578 pKa = 4.12QIDD581 pKa = 3.41ISSSGSEE588 pKa = 3.86TSAKK592 pKa = 8.72QTTTEE597 pKa = 4.53TIPDD601 pKa = 3.42NHH603 pKa = 6.29TTDD606 pKa = 4.0IVQEE610 pKa = 4.21DD611 pKa = 4.52PGTEE615 pKa = 3.64ILSVEE620 pKa = 4.19NTTVRR625 pKa = 11.84VPDD628 pKa = 4.85DD629 pKa = 3.67NTTDD633 pKa = 3.27ISPSDD638 pKa = 3.85VICNSEE644 pKa = 4.1NEE646 pKa = 4.33ASSEE650 pKa = 4.01PNEE653 pKa = 3.71TRR655 pKa = 11.84TYY657 pKa = 10.59VSIVPTTKK665 pKa = 10.63SKK667 pKa = 11.36DD668 pKa = 3.5STITSAFSGYY678 pKa = 9.74EE679 pKa = 3.27QDD681 pKa = 4.15TITKK685 pKa = 9.5VITYY689 pKa = 7.28TTTICEE695 pKa = 4.51SDD697 pKa = 3.46TVTQVITYY705 pKa = 6.74TTIITNHH712 pKa = 6.63DD713 pKa = 4.59SIAKK717 pKa = 7.96TATEE721 pKa = 4.23MTTEE725 pKa = 3.99ATSFAVTTVGEE736 pKa = 4.34TRR738 pKa = 11.84TIIYY742 pKa = 9.54LQDD745 pKa = 3.32SMGTTNDD752 pKa = 3.15TGNNSSNTKK761 pKa = 9.86SDD763 pKa = 2.98SRR765 pKa = 11.84TASNNTGNNSSNTKK779 pKa = 9.92SYY781 pKa = 11.19SGTASNEE788 pKa = 4.14EE789 pKa = 3.93IQTTANSEE797 pKa = 4.18NNSEE801 pKa = 4.22HH802 pKa = 6.07TKK804 pKa = 10.56SEE806 pKa = 4.14SGQSDD811 pKa = 2.61ISEE814 pKa = 4.49YY815 pKa = 10.86YY816 pKa = 10.52SGGTEE821 pKa = 3.9SDD823 pKa = 3.19SGKK826 pKa = 10.47GNKK829 pKa = 9.52SVVPEE834 pKa = 4.16EE835 pKa = 4.09NQGTNSRR842 pKa = 11.84NNNSSTNSNNRR853 pKa = 11.84SDD855 pKa = 4.41DD856 pKa = 3.26QTTRR860 pKa = 11.84GSYY863 pKa = 8.49STSLQLVTSNGIGSTRR879 pKa = 11.84GTSTFSLLFTFVIAILYY896 pKa = 8.94
Molecular weight: 97.34 kDa
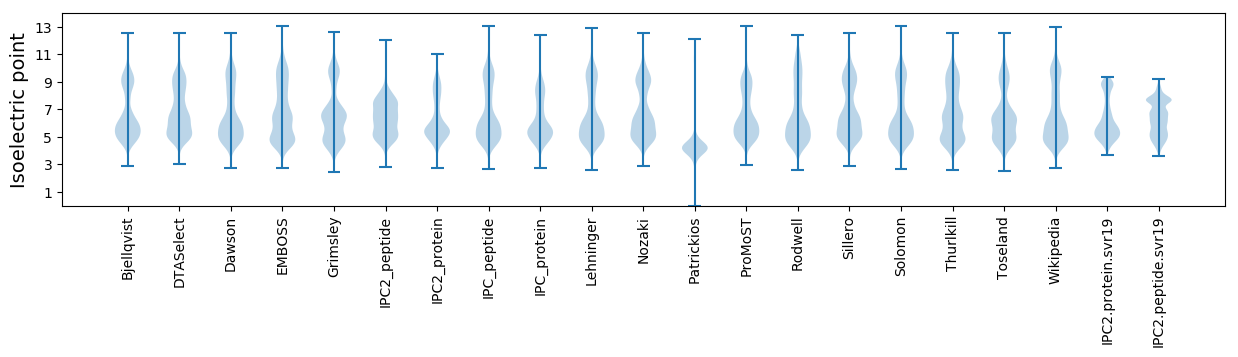
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G3ATQ7|G3ATQ7_SPAPN WD_REPEATS_REGION domain-containing protein OS=Spathaspora passalidarum (strain NRRL Y-27907 / 11-Y1) OX=619300 GN=SPAPADRAFT_52383 PE=3 SV=1
MM1 pKa = 7.94PSQKK5 pKa = 10.41SFRR8 pKa = 11.84TKK10 pKa = 10.38QKK12 pKa = 9.84LAKK15 pKa = 9.55AQKK18 pKa = 9.21QNRR21 pKa = 11.84PLPQWVRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.88LGII51 pKa = 4.0
MM1 pKa = 7.94PSQKK5 pKa = 10.41SFRR8 pKa = 11.84TKK10 pKa = 10.38QKK12 pKa = 9.84LAKK15 pKa = 9.55AQKK18 pKa = 9.21QNRR21 pKa = 11.84PLPQWVRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.88LGII51 pKa = 4.0
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2693134 |
49 |
4210 |
450.9 |
50.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.585 ± 0.028 | 1.11 ± 0.01 |
5.896 ± 0.025 | 6.759 ± 0.039 |
4.453 ± 0.019 | 5.219 ± 0.033 |
2.212 ± 0.014 | 6.926 ± 0.022 |
6.96 ± 0.033 | 9.351 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.9 ± 0.011 | 5.599 ± 0.025 |
4.68 ± 0.028 | 4.269 ± 0.032 |
4.115 ± 0.021 | 8.434 ± 0.032 |
5.96 ± 0.034 | 5.929 ± 0.024 |
1.025 ± 0.01 | 3.617 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
