
Carnation mottle virus (isolate China/Shanghai) (CarMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Alphacarmovirus; Carnation mottle virus
Average proteome isoelectric point is 8.49
Get precalculated fractions of proteins
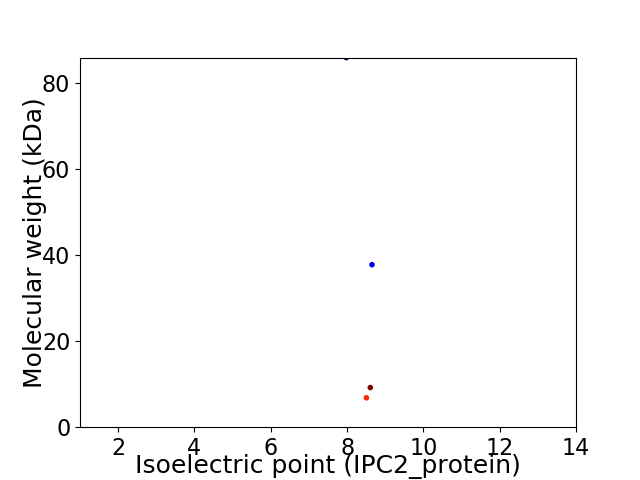
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|Q9Q6X7|CAPSD_CARMS Capsid protein OS=Carnation mottle virus (isolate China/Shanghai) OX=652111 GN=ORF4 PE=3 SV=1
MM1 pKa = 7.72GLPSLLVEE9 pKa = 4.26GVIGCTLVGGLVAVGSAALAVRR31 pKa = 11.84ATIGVVEE38 pKa = 5.01FNRR41 pKa = 11.84EE42 pKa = 4.02CVRR45 pKa = 11.84GARR48 pKa = 11.84RR49 pKa = 11.84IVSSGGRR56 pKa = 11.84CLIVQSPYY64 pKa = 11.2GNPNQGLIRR73 pKa = 11.84GEE75 pKa = 4.14DD76 pKa = 3.53EE77 pKa = 5.18EE78 pKa = 4.91IDD80 pKa = 3.61NVEE83 pKa = 3.81EE84 pKa = 4.23STPEE88 pKa = 3.88EE89 pKa = 4.0LTQLIEE95 pKa = 4.22VKK97 pKa = 10.96AEE99 pKa = 3.77VDD101 pKa = 3.33GKK103 pKa = 10.75EE104 pKa = 3.92VVVSKK109 pKa = 10.81KK110 pKa = 10.33RR111 pKa = 11.84VVNRR115 pKa = 11.84HH116 pKa = 3.72LRR118 pKa = 11.84QRR120 pKa = 11.84FVRR123 pKa = 11.84SIAIEE128 pKa = 4.13AKK130 pKa = 9.42NHH132 pKa = 6.32FGGDD136 pKa = 3.37ISPSKK141 pKa = 11.03ANYY144 pKa = 9.88LSVSKK149 pKa = 10.47FLTGKK154 pKa = 9.73CKK156 pKa = 9.81EE157 pKa = 4.08RR158 pKa = 11.84HH159 pKa = 5.0VVPAHH164 pKa = 5.86TRR166 pKa = 11.84DD167 pKa = 3.79CVSAAMVLVFTPDD180 pKa = 2.82VHH182 pKa = 7.06EE183 pKa = 4.17IRR185 pKa = 11.84MMAGLASDD193 pKa = 3.85AAYY196 pKa = 10.46ARR198 pKa = 11.84KK199 pKa = 9.2IAMASILNRR208 pKa = 11.84KK209 pKa = 6.94GWCWRR214 pKa = 11.84LMVNPLDD221 pKa = 3.81RR222 pKa = 11.84ARR224 pKa = 11.84WWEE227 pKa = 3.83MWCVVNGFDD236 pKa = 4.61SNKK239 pKa = 9.88PVTFPKK245 pKa = 10.05XGGLFYY251 pKa = 11.2LNGVEE256 pKa = 4.25TKK258 pKa = 9.69IRR260 pKa = 11.84RR261 pKa = 11.84GGHH264 pKa = 5.7PSVIEE269 pKa = 4.06VDD271 pKa = 4.24GQCPPRR277 pKa = 11.84EE278 pKa = 3.76RR279 pKa = 11.84KK280 pKa = 10.11LYY282 pKa = 8.08VQNAITTGYY291 pKa = 9.48EE292 pKa = 4.01YY293 pKa = 10.37RR294 pKa = 11.84VHH296 pKa = 5.53NHH298 pKa = 5.51SYY300 pKa = 10.18ANLRR304 pKa = 11.84RR305 pKa = 11.84GLLEE309 pKa = 3.45RR310 pKa = 11.84VFYY313 pKa = 10.17VEE315 pKa = 4.06RR316 pKa = 11.84NKK318 pKa = 10.85EE319 pKa = 4.13LVSCPQPEE327 pKa = 4.1PGSFKK332 pKa = 10.84EE333 pKa = 3.58MGYY336 pKa = 10.1LRR338 pKa = 11.84RR339 pKa = 11.84RR340 pKa = 11.84FHH342 pKa = 6.55RR343 pKa = 11.84VCGNHH348 pKa = 5.21TRR350 pKa = 11.84ISANDD355 pKa = 3.74LVDD358 pKa = 4.53CYY360 pKa = 10.57QGRR363 pKa = 11.84KK364 pKa = 6.25RR365 pKa = 11.84TIYY368 pKa = 10.23EE369 pKa = 3.57NAAASLLDD377 pKa = 3.58RR378 pKa = 11.84AIEE381 pKa = 4.34RR382 pKa = 11.84KK383 pKa = 10.47DD384 pKa = 3.29GDD386 pKa = 3.85LKK388 pKa = 10.58TFIKK392 pKa = 10.72AEE394 pKa = 4.03KK395 pKa = 10.29FNVNLKK401 pKa = 10.13SDD403 pKa = 3.86PAPRR407 pKa = 11.84VIQPRR412 pKa = 11.84SPRR415 pKa = 11.84YY416 pKa = 8.45NVEE419 pKa = 3.57LGRR422 pKa = 11.84YY423 pKa = 6.83LKK425 pKa = 10.42KK426 pKa = 10.69YY427 pKa = 7.28EE428 pKa = 3.77HH429 pKa = 6.89HH430 pKa = 7.16AYY432 pKa = 9.6KK433 pKa = 10.64ALDD436 pKa = 4.48KK437 pKa = 10.56IWGGPTVMKK446 pKa = 10.54GYY448 pKa = 6.47TTEE451 pKa = 4.47EE452 pKa = 4.26VAQHH456 pKa = 6.32IWSAWNQFQTPVAIGFDD473 pKa = 3.33MSRR476 pKa = 11.84FDD478 pKa = 3.3QHH480 pKa = 7.95VSVAALEE487 pKa = 4.71FEE489 pKa = 4.49HH490 pKa = 6.98SCYY493 pKa = 10.15LACFEE498 pKa = 5.13GDD500 pKa = 2.96AHH502 pKa = 7.19LANLLKK508 pKa = 9.92MQLVNHH514 pKa = 6.39GVGFASNGMLRR525 pKa = 11.84YY526 pKa = 7.8TKK528 pKa = 9.9EE529 pKa = 3.9GCRR532 pKa = 11.84MIGDD536 pKa = 3.9MNTALGNCLLACLITKK552 pKa = 9.93HH553 pKa = 5.65LMKK556 pKa = 10.23IRR558 pKa = 11.84SRR560 pKa = 11.84LINNGDD566 pKa = 3.76DD567 pKa = 3.44CVLICEE573 pKa = 4.63RR574 pKa = 11.84TDD576 pKa = 2.74IDD578 pKa = 4.14YY579 pKa = 11.16VVSNLTTGWSRR590 pKa = 11.84FGFNCIAEE598 pKa = 4.26EE599 pKa = 3.86PVYY602 pKa = 10.64EE603 pKa = 4.01MEE605 pKa = 6.04KK606 pKa = 10.26IRR608 pKa = 11.84FCQMAPVFDD617 pKa = 3.99GAGWLMVRR625 pKa = 11.84DD626 pKa = 4.39PLVSMSKK633 pKa = 10.33DD634 pKa = 3.3SHH636 pKa = 8.07SLVHH640 pKa = 6.49WNNEE644 pKa = 3.86TNAKK648 pKa = 9.0QWLKK652 pKa = 10.83SVGMCGLRR660 pKa = 11.84IAGGVPIVQEE670 pKa = 4.92FYY672 pKa = 10.73QKK674 pKa = 10.73YY675 pKa = 9.0VEE677 pKa = 4.14TAGNVKK683 pKa = 9.75EE684 pKa = 4.45NKK686 pKa = 10.23NITEE690 pKa = 4.16KK691 pKa = 10.73SSSGFFMMADD701 pKa = 3.53RR702 pKa = 11.84AKK704 pKa = 10.04RR705 pKa = 11.84GYY707 pKa = 9.89SAVSEE712 pKa = 4.09MCRR715 pKa = 11.84FSFYY719 pKa = 10.43QAFGITPDD727 pKa = 3.41QQIALEE733 pKa = 4.18GEE735 pKa = 4.21IRR737 pKa = 11.84SLTIDD742 pKa = 3.63TNVGPQCEE750 pKa = 4.32AADD753 pKa = 4.06SLWILNRR760 pKa = 11.84KK761 pKa = 6.89YY762 pKa = 10.03QQ763 pKa = 3.42
MM1 pKa = 7.72GLPSLLVEE9 pKa = 4.26GVIGCTLVGGLVAVGSAALAVRR31 pKa = 11.84ATIGVVEE38 pKa = 5.01FNRR41 pKa = 11.84EE42 pKa = 4.02CVRR45 pKa = 11.84GARR48 pKa = 11.84RR49 pKa = 11.84IVSSGGRR56 pKa = 11.84CLIVQSPYY64 pKa = 11.2GNPNQGLIRR73 pKa = 11.84GEE75 pKa = 4.14DD76 pKa = 3.53EE77 pKa = 5.18EE78 pKa = 4.91IDD80 pKa = 3.61NVEE83 pKa = 3.81EE84 pKa = 4.23STPEE88 pKa = 3.88EE89 pKa = 4.0LTQLIEE95 pKa = 4.22VKK97 pKa = 10.96AEE99 pKa = 3.77VDD101 pKa = 3.33GKK103 pKa = 10.75EE104 pKa = 3.92VVVSKK109 pKa = 10.81KK110 pKa = 10.33RR111 pKa = 11.84VVNRR115 pKa = 11.84HH116 pKa = 3.72LRR118 pKa = 11.84QRR120 pKa = 11.84FVRR123 pKa = 11.84SIAIEE128 pKa = 4.13AKK130 pKa = 9.42NHH132 pKa = 6.32FGGDD136 pKa = 3.37ISPSKK141 pKa = 11.03ANYY144 pKa = 9.88LSVSKK149 pKa = 10.47FLTGKK154 pKa = 9.73CKK156 pKa = 9.81EE157 pKa = 4.08RR158 pKa = 11.84HH159 pKa = 5.0VVPAHH164 pKa = 5.86TRR166 pKa = 11.84DD167 pKa = 3.79CVSAAMVLVFTPDD180 pKa = 2.82VHH182 pKa = 7.06EE183 pKa = 4.17IRR185 pKa = 11.84MMAGLASDD193 pKa = 3.85AAYY196 pKa = 10.46ARR198 pKa = 11.84KK199 pKa = 9.2IAMASILNRR208 pKa = 11.84KK209 pKa = 6.94GWCWRR214 pKa = 11.84LMVNPLDD221 pKa = 3.81RR222 pKa = 11.84ARR224 pKa = 11.84WWEE227 pKa = 3.83MWCVVNGFDD236 pKa = 4.61SNKK239 pKa = 9.88PVTFPKK245 pKa = 10.05XGGLFYY251 pKa = 11.2LNGVEE256 pKa = 4.25TKK258 pKa = 9.69IRR260 pKa = 11.84RR261 pKa = 11.84GGHH264 pKa = 5.7PSVIEE269 pKa = 4.06VDD271 pKa = 4.24GQCPPRR277 pKa = 11.84EE278 pKa = 3.76RR279 pKa = 11.84KK280 pKa = 10.11LYY282 pKa = 8.08VQNAITTGYY291 pKa = 9.48EE292 pKa = 4.01YY293 pKa = 10.37RR294 pKa = 11.84VHH296 pKa = 5.53NHH298 pKa = 5.51SYY300 pKa = 10.18ANLRR304 pKa = 11.84RR305 pKa = 11.84GLLEE309 pKa = 3.45RR310 pKa = 11.84VFYY313 pKa = 10.17VEE315 pKa = 4.06RR316 pKa = 11.84NKK318 pKa = 10.85EE319 pKa = 4.13LVSCPQPEE327 pKa = 4.1PGSFKK332 pKa = 10.84EE333 pKa = 3.58MGYY336 pKa = 10.1LRR338 pKa = 11.84RR339 pKa = 11.84RR340 pKa = 11.84FHH342 pKa = 6.55RR343 pKa = 11.84VCGNHH348 pKa = 5.21TRR350 pKa = 11.84ISANDD355 pKa = 3.74LVDD358 pKa = 4.53CYY360 pKa = 10.57QGRR363 pKa = 11.84KK364 pKa = 6.25RR365 pKa = 11.84TIYY368 pKa = 10.23EE369 pKa = 3.57NAAASLLDD377 pKa = 3.58RR378 pKa = 11.84AIEE381 pKa = 4.34RR382 pKa = 11.84KK383 pKa = 10.47DD384 pKa = 3.29GDD386 pKa = 3.85LKK388 pKa = 10.58TFIKK392 pKa = 10.72AEE394 pKa = 4.03KK395 pKa = 10.29FNVNLKK401 pKa = 10.13SDD403 pKa = 3.86PAPRR407 pKa = 11.84VIQPRR412 pKa = 11.84SPRR415 pKa = 11.84YY416 pKa = 8.45NVEE419 pKa = 3.57LGRR422 pKa = 11.84YY423 pKa = 6.83LKK425 pKa = 10.42KK426 pKa = 10.69YY427 pKa = 7.28EE428 pKa = 3.77HH429 pKa = 6.89HH430 pKa = 7.16AYY432 pKa = 9.6KK433 pKa = 10.64ALDD436 pKa = 4.48KK437 pKa = 10.56IWGGPTVMKK446 pKa = 10.54GYY448 pKa = 6.47TTEE451 pKa = 4.47EE452 pKa = 4.26VAQHH456 pKa = 6.32IWSAWNQFQTPVAIGFDD473 pKa = 3.33MSRR476 pKa = 11.84FDD478 pKa = 3.3QHH480 pKa = 7.95VSVAALEE487 pKa = 4.71FEE489 pKa = 4.49HH490 pKa = 6.98SCYY493 pKa = 10.15LACFEE498 pKa = 5.13GDD500 pKa = 2.96AHH502 pKa = 7.19LANLLKK508 pKa = 9.92MQLVNHH514 pKa = 6.39GVGFASNGMLRR525 pKa = 11.84YY526 pKa = 7.8TKK528 pKa = 9.9EE529 pKa = 3.9GCRR532 pKa = 11.84MIGDD536 pKa = 3.9MNTALGNCLLACLITKK552 pKa = 9.93HH553 pKa = 5.65LMKK556 pKa = 10.23IRR558 pKa = 11.84SRR560 pKa = 11.84LINNGDD566 pKa = 3.76DD567 pKa = 3.44CVLICEE573 pKa = 4.63RR574 pKa = 11.84TDD576 pKa = 2.74IDD578 pKa = 4.14YY579 pKa = 11.16VVSNLTTGWSRR590 pKa = 11.84FGFNCIAEE598 pKa = 4.26EE599 pKa = 3.86PVYY602 pKa = 10.64EE603 pKa = 4.01MEE605 pKa = 6.04KK606 pKa = 10.26IRR608 pKa = 11.84FCQMAPVFDD617 pKa = 3.99GAGWLMVRR625 pKa = 11.84DD626 pKa = 4.39PLVSMSKK633 pKa = 10.33DD634 pKa = 3.3SHH636 pKa = 8.07SLVHH640 pKa = 6.49WNNEE644 pKa = 3.86TNAKK648 pKa = 9.0QWLKK652 pKa = 10.83SVGMCGLRR660 pKa = 11.84IAGGVPIVQEE670 pKa = 4.92FYY672 pKa = 10.73QKK674 pKa = 10.73YY675 pKa = 9.0VEE677 pKa = 4.14TAGNVKK683 pKa = 9.75EE684 pKa = 4.45NKK686 pKa = 10.23NITEE690 pKa = 4.16KK691 pKa = 10.73SSSGFFMMADD701 pKa = 3.53RR702 pKa = 11.84AKK704 pKa = 10.04RR705 pKa = 11.84GYY707 pKa = 9.89SAVSEE712 pKa = 4.09MCRR715 pKa = 11.84FSFYY719 pKa = 10.43QAFGITPDD727 pKa = 3.41QQIALEE733 pKa = 4.18GEE735 pKa = 4.21IRR737 pKa = 11.84SLTIDD742 pKa = 3.63TNVGPQCEE750 pKa = 4.32AADD753 pKa = 4.06SLWILNRR760 pKa = 11.84KK761 pKa = 6.89YY762 pKa = 10.03QQ763 pKa = 3.42
Molecular weight: 85.85 kDa
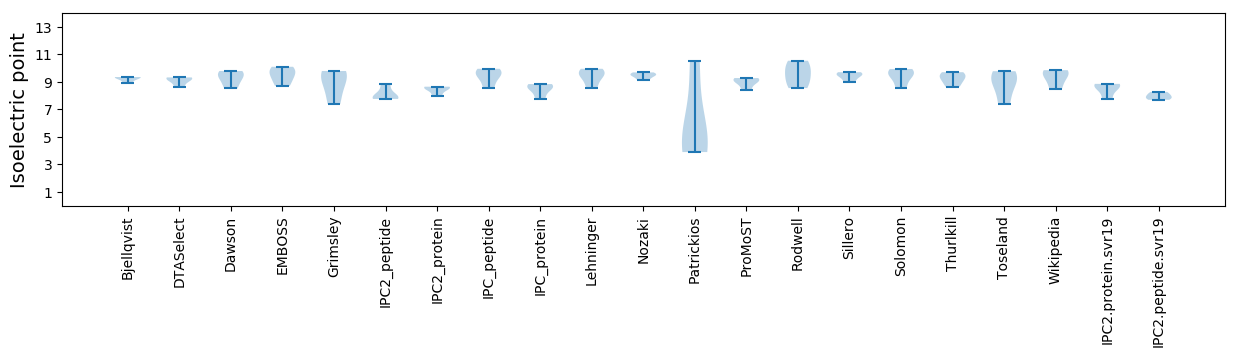
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9J5U7|RDRP_CARMS Probable RNA-directed RNA polymerase OS=Carnation mottle virus (isolate China/Shanghai) OX=652111 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.77PSANLHH7 pKa = 6.35PIVLTGVIGLMLLIRR22 pKa = 11.84LRR24 pKa = 11.84CTFTSTFSLPPLVTLNQIIALSFCGLLLNSISRR57 pKa = 11.84AEE59 pKa = 3.86RR60 pKa = 11.84ACYY63 pKa = 10.16YY64 pKa = 10.23NYY66 pKa = 10.88SVDD69 pKa = 3.47SSKK72 pKa = 10.36QQHH75 pKa = 6.02ISISTPNGKK84 pKa = 9.32
MM1 pKa = 7.77PSANLHH7 pKa = 6.35PIVLTGVIGLMLLIRR22 pKa = 11.84LRR24 pKa = 11.84CTFTSTFSLPPLVTLNQIIALSFCGLLLNSISRR57 pKa = 11.84AEE59 pKa = 3.86RR60 pKa = 11.84ACYY63 pKa = 10.16YY64 pKa = 10.23NYY66 pKa = 10.88SVDD69 pKa = 3.47SSKK72 pKa = 10.36QQHH75 pKa = 6.02ISISTPNGKK84 pKa = 9.32
Molecular weight: 9.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1256 |
61 |
763 |
314.0 |
34.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.404 ± 0.436 | 2.389 ± 0.621 |
4.459 ± 0.588 | 5.573 ± 1.08 |
3.662 ± 0.126 | 7.484 ± 0.587 |
1.99 ± 0.668 | 5.414 ± 0.817 |
5.971 ± 0.78 | 7.803 ± 1.252 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.866 ± 0.148 | 5.016 ± 0.716 |
4.538 ± 0.742 | 3.185 ± 0.15 |
6.608 ± 0.749 | 6.608 ± 1.101 |
5.812 ± 1.729 | 8.758 ± 0.649 |
1.354 ± 0.338 | 3.025 ± 0.529 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
