
Faeces associated gemycircularvirus 15
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemygorvirus; Gemygorvirus cania1; Canine associated gemygorvirus 1
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
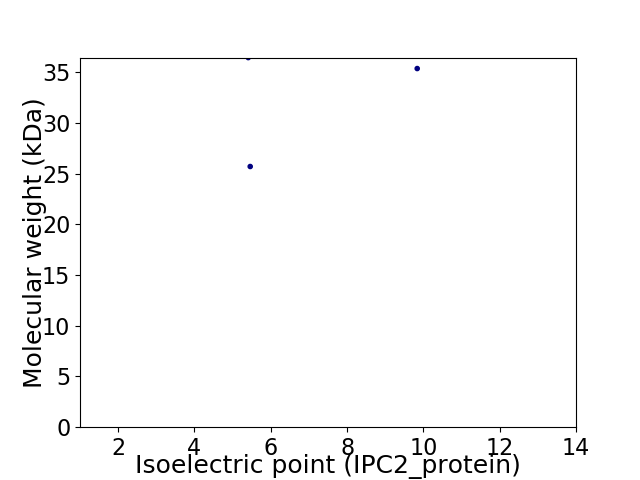
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MGD9|A0A168MGD9_9VIRU Replication-associated protein OS=Faeces associated gemycircularvirus 15 OX=1843735 PE=3 SV=1
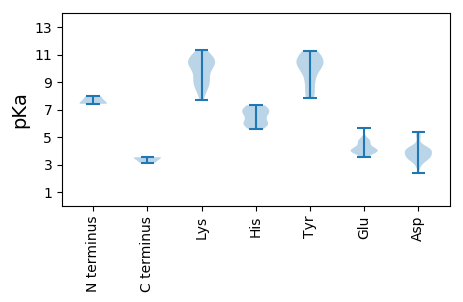
MM1 pKa = 7.42SNSFRR6 pKa = 11.84LQARR10 pKa = 11.84YY11 pKa = 10.17VLLTYY16 pKa = 8.05SQCGDD21 pKa = 3.75LDD23 pKa = 3.26PWMVCDD29 pKa = 5.19HH30 pKa = 6.72LASLHH35 pKa = 6.02AEE37 pKa = 3.92CLIGRR42 pKa = 11.84EE43 pKa = 3.97NHH45 pKa = 5.59VDD47 pKa = 3.5GGIHH51 pKa = 6.05LHH53 pKa = 6.64AFVDD57 pKa = 4.11LGKK60 pKa = 9.14KK61 pKa = 8.32TDD63 pKa = 3.36IRR65 pKa = 11.84DD66 pKa = 3.35SRR68 pKa = 11.84LFDD71 pKa = 4.2VNGQHH76 pKa = 7.32PNITPCGRR84 pKa = 11.84TPQKK88 pKa = 10.31MLDD91 pKa = 3.71YY92 pKa = 10.33AVKK95 pKa = 10.78DD96 pKa = 3.61GDD98 pKa = 4.12VVAGGLDD105 pKa = 3.47PVLGDD110 pKa = 3.96EE111 pKa = 4.61VSSADD116 pKa = 4.15TVWHH120 pKa = 6.89RR121 pKa = 11.84IANAATVDD129 pKa = 4.08EE130 pKa = 4.85FWVITRR136 pKa = 11.84EE137 pKa = 3.96LAPRR141 pKa = 11.84ALLCNHH147 pKa = 5.94QSLRR151 pKa = 11.84AYY153 pKa = 10.25AEE155 pKa = 3.53WHH157 pKa = 5.77YY158 pKa = 11.06RR159 pKa = 11.84PPTLVYY165 pKa = 9.03EE166 pKa = 4.78HH167 pKa = 7.17PCEE170 pKa = 3.95LRR172 pKa = 11.84LRR174 pKa = 11.84ADD176 pKa = 4.23RR177 pKa = 11.84LPEE180 pKa = 3.99LDD182 pKa = 2.91EE183 pKa = 4.04WVRR186 pKa = 11.84EE187 pKa = 4.23SLSGTGGRR195 pKa = 11.84PKK197 pKa = 10.75SLILWGEE204 pKa = 4.09SKK206 pKa = 10.67LGKK209 pKa = 8.69TIWARR214 pKa = 11.84SLGQHH219 pKa = 6.94IYY221 pKa = 11.12CCMQFNVDD229 pKa = 4.01DD230 pKa = 3.95VRR232 pKa = 11.84ANLQEE237 pKa = 3.82AQYY240 pKa = 11.22AIFDD244 pKa = 5.01DD245 pKa = 3.84IQGNFQFFPSYY256 pKa = 10.36KK257 pKa = 9.0GWLGAQHH264 pKa = 6.28QFTVTDD270 pKa = 3.81KK271 pKa = 11.3YY272 pKa = 10.99KK273 pKa = 11.19GKK275 pKa = 7.67TTINWGRR282 pKa = 11.84PSIWLTNDD290 pKa = 2.94DD291 pKa = 4.27PEE293 pKa = 4.58EE294 pKa = 4.24VGHH297 pKa = 7.26VDD299 pKa = 5.37LNWLRR304 pKa = 11.84KK305 pKa = 9.22NCVIVNITEE314 pKa = 4.55SIVEE318 pKa = 4.04LVV320 pKa = 3.13
MM1 pKa = 7.42SNSFRR6 pKa = 11.84LQARR10 pKa = 11.84YY11 pKa = 10.17VLLTYY16 pKa = 8.05SQCGDD21 pKa = 3.75LDD23 pKa = 3.26PWMVCDD29 pKa = 5.19HH30 pKa = 6.72LASLHH35 pKa = 6.02AEE37 pKa = 3.92CLIGRR42 pKa = 11.84EE43 pKa = 3.97NHH45 pKa = 5.59VDD47 pKa = 3.5GGIHH51 pKa = 6.05LHH53 pKa = 6.64AFVDD57 pKa = 4.11LGKK60 pKa = 9.14KK61 pKa = 8.32TDD63 pKa = 3.36IRR65 pKa = 11.84DD66 pKa = 3.35SRR68 pKa = 11.84LFDD71 pKa = 4.2VNGQHH76 pKa = 7.32PNITPCGRR84 pKa = 11.84TPQKK88 pKa = 10.31MLDD91 pKa = 3.71YY92 pKa = 10.33AVKK95 pKa = 10.78DD96 pKa = 3.61GDD98 pKa = 4.12VVAGGLDD105 pKa = 3.47PVLGDD110 pKa = 3.96EE111 pKa = 4.61VSSADD116 pKa = 4.15TVWHH120 pKa = 6.89RR121 pKa = 11.84IANAATVDD129 pKa = 4.08EE130 pKa = 4.85FWVITRR136 pKa = 11.84EE137 pKa = 3.96LAPRR141 pKa = 11.84ALLCNHH147 pKa = 5.94QSLRR151 pKa = 11.84AYY153 pKa = 10.25AEE155 pKa = 3.53WHH157 pKa = 5.77YY158 pKa = 11.06RR159 pKa = 11.84PPTLVYY165 pKa = 9.03EE166 pKa = 4.78HH167 pKa = 7.17PCEE170 pKa = 3.95LRR172 pKa = 11.84LRR174 pKa = 11.84ADD176 pKa = 4.23RR177 pKa = 11.84LPEE180 pKa = 3.99LDD182 pKa = 2.91EE183 pKa = 4.04WVRR186 pKa = 11.84EE187 pKa = 4.23SLSGTGGRR195 pKa = 11.84PKK197 pKa = 10.75SLILWGEE204 pKa = 4.09SKK206 pKa = 10.67LGKK209 pKa = 8.69TIWARR214 pKa = 11.84SLGQHH219 pKa = 6.94IYY221 pKa = 11.12CCMQFNVDD229 pKa = 4.01DD230 pKa = 3.95VRR232 pKa = 11.84ANLQEE237 pKa = 3.82AQYY240 pKa = 11.22AIFDD244 pKa = 5.01DD245 pKa = 3.84IQGNFQFFPSYY256 pKa = 10.36KK257 pKa = 9.0GWLGAQHH264 pKa = 6.28QFTVTDD270 pKa = 3.81KK271 pKa = 11.3YY272 pKa = 10.99KK273 pKa = 11.19GKK275 pKa = 7.67TTINWGRR282 pKa = 11.84PSIWLTNDD290 pKa = 2.94DD291 pKa = 4.27PEE293 pKa = 4.58EE294 pKa = 4.24VGHH297 pKa = 7.26VDD299 pKa = 5.37LNWLRR304 pKa = 11.84KK305 pKa = 9.22NCVIVNITEE314 pKa = 4.55SIVEE318 pKa = 4.04LVV320 pKa = 3.13
Molecular weight: 36.43 kDa
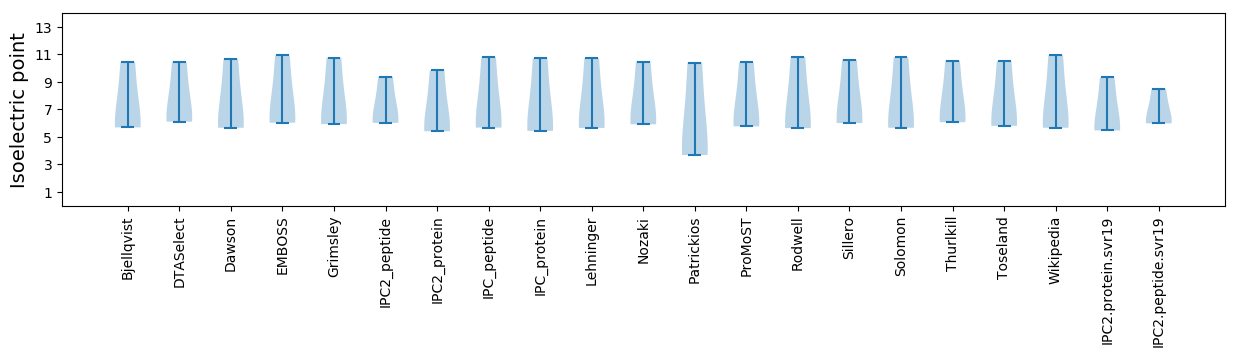
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161IBC4|A0A161IBC4_9VIRU RepA OS=Faeces associated gemycircularvirus 15 OX=1843735 PE=3 SV=1
MM1 pKa = 7.97PIRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SSRR9 pKa = 11.84RR10 pKa = 11.84SAGKK14 pKa = 8.84RR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 8.08PRR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.25RR22 pKa = 11.84KK23 pKa = 9.91APARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84QTRR33 pKa = 11.84RR34 pKa = 11.84MPARR38 pKa = 11.84RR39 pKa = 11.84VTRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ILNVSSIKK52 pKa = 10.52KK53 pKa = 8.93RR54 pKa = 11.84DD55 pKa = 3.48NMMSAVINPGEE66 pKa = 4.1ARR68 pKa = 11.84PTLGPLTTTTGFTSLYY84 pKa = 9.72IPTARR89 pKa = 11.84TLAPANDD96 pKa = 3.65QGEE99 pKa = 4.78SMRR102 pKa = 11.84QRR104 pKa = 11.84QTTYY108 pKa = 10.75AGWVQRR114 pKa = 11.84AVQVDD119 pKa = 4.18IIGGGVWKK127 pKa = 9.79WRR129 pKa = 11.84RR130 pKa = 11.84IVFAYY135 pKa = 9.75KK136 pKa = 10.36GGEE139 pKa = 4.23SLWTGEE145 pKa = 5.67FPGQWTEE152 pKa = 3.79PFFSKK157 pKa = 10.32SVKK160 pKa = 9.75PDD162 pKa = 3.67PDD164 pKa = 4.33DD165 pKa = 4.63PLPIAPDD172 pKa = 3.39MVRR175 pKa = 11.84LIGQPQSAQANAIRR189 pKa = 11.84DD190 pKa = 4.09LLWDD194 pKa = 3.39GHH196 pKa = 6.32EE197 pKa = 4.33GVDD200 pKa = 3.76WSSQYY205 pKa = 7.83TAKK208 pKa = 10.54VDD210 pKa = 3.68TKK212 pKa = 10.35RR213 pKa = 11.84VRR215 pKa = 11.84LMSDD219 pKa = 2.37KK220 pKa = 10.76TYY222 pKa = 11.18VFNPGNEE229 pKa = 4.15SGMSRR234 pKa = 11.84TYY236 pKa = 10.76RR237 pKa = 11.84FWYY240 pKa = 9.11PIGHH244 pKa = 6.73NIVYY248 pKa = 10.56DD249 pKa = 3.98DD250 pKa = 5.03DD251 pKa = 3.79EE252 pKa = 4.65WGASAFGRR260 pKa = 11.84VSPISVQSKK269 pKa = 10.21LGFGDD274 pKa = 3.57VYY276 pKa = 10.65IYY278 pKa = 10.41DD279 pKa = 3.18IAYY282 pKa = 9.65RR283 pKa = 11.84VVPSSGSVEE292 pKa = 3.81AQMQFSPEE300 pKa = 3.53GTYY303 pKa = 10.33YY304 pKa = 8.87WHH306 pKa = 6.98EE307 pKa = 4.02RR308 pKa = 3.5
MM1 pKa = 7.97PIRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SSRR9 pKa = 11.84RR10 pKa = 11.84SAGKK14 pKa = 8.84RR15 pKa = 11.84RR16 pKa = 11.84YY17 pKa = 8.08PRR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.25RR22 pKa = 11.84KK23 pKa = 9.91APARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84QTRR33 pKa = 11.84RR34 pKa = 11.84MPARR38 pKa = 11.84RR39 pKa = 11.84VTRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ILNVSSIKK52 pKa = 10.52KK53 pKa = 8.93RR54 pKa = 11.84DD55 pKa = 3.48NMMSAVINPGEE66 pKa = 4.1ARR68 pKa = 11.84PTLGPLTTTTGFTSLYY84 pKa = 9.72IPTARR89 pKa = 11.84TLAPANDD96 pKa = 3.65QGEE99 pKa = 4.78SMRR102 pKa = 11.84QRR104 pKa = 11.84QTTYY108 pKa = 10.75AGWVQRR114 pKa = 11.84AVQVDD119 pKa = 4.18IIGGGVWKK127 pKa = 9.79WRR129 pKa = 11.84RR130 pKa = 11.84IVFAYY135 pKa = 9.75KK136 pKa = 10.36GGEE139 pKa = 4.23SLWTGEE145 pKa = 5.67FPGQWTEE152 pKa = 3.79PFFSKK157 pKa = 10.32SVKK160 pKa = 9.75PDD162 pKa = 3.67PDD164 pKa = 4.33DD165 pKa = 4.63PLPIAPDD172 pKa = 3.39MVRR175 pKa = 11.84LIGQPQSAQANAIRR189 pKa = 11.84DD190 pKa = 4.09LLWDD194 pKa = 3.39GHH196 pKa = 6.32EE197 pKa = 4.33GVDD200 pKa = 3.76WSSQYY205 pKa = 7.83TAKK208 pKa = 10.54VDD210 pKa = 3.68TKK212 pKa = 10.35RR213 pKa = 11.84VRR215 pKa = 11.84LMSDD219 pKa = 2.37KK220 pKa = 10.76TYY222 pKa = 11.18VFNPGNEE229 pKa = 4.15SGMSRR234 pKa = 11.84TYY236 pKa = 10.76RR237 pKa = 11.84FWYY240 pKa = 9.11PIGHH244 pKa = 6.73NIVYY248 pKa = 10.56DD249 pKa = 3.98DD250 pKa = 5.03DD251 pKa = 3.79EE252 pKa = 4.65WGASAFGRR260 pKa = 11.84VSPISVQSKK269 pKa = 10.21LGFGDD274 pKa = 3.57VYY276 pKa = 10.65IYY278 pKa = 10.41DD279 pKa = 3.18IAYY282 pKa = 9.65RR283 pKa = 11.84VVPSSGSVEE292 pKa = 3.81AQMQFSPEE300 pKa = 3.53GTYY303 pKa = 10.33YY304 pKa = 8.87WHH306 pKa = 6.98EE307 pKa = 4.02RR308 pKa = 3.5
Molecular weight: 35.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861 |
233 |
320 |
287.0 |
32.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.433 ± 0.752 | 2.091 ± 0.933 |
6.969 ± 0.647 | 4.53 ± 0.455 |
2.904 ± 0.237 | 7.666 ± 0.238 |
3.02 ± 0.907 | 4.413 ± 0.569 |
3.252 ± 0.475 | 7.898 ± 1.793 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.974 ± 0.442 | 3.252 ± 0.489 |
5.575 ± 0.731 | 3.833 ± 0.413 |
8.827 ± 1.621 | 6.736 ± 0.807 |
5.575 ± 0.3 | 7.201 ± 0.179 |
3.136 ± 0.176 | 3.717 ± 0.512 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
