
Sclerotinia sclerotiorum mitovirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H2EQQ4|H2EQQ4_9VIRU RNA-dependent RNA polymerases OS=Sclerotinia sclerotiorum mitovirus 2 OX=1133728 GN=SsMV2_gp1 PE=4 SV=1
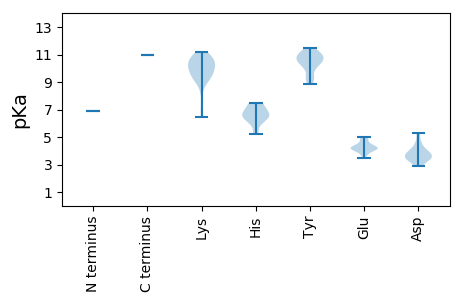
MM1 pKa = 6.89TQPLVLKK8 pKa = 10.31LMKK11 pKa = 9.52WVRR14 pKa = 11.84KK15 pKa = 9.24YY16 pKa = 10.61YY17 pKa = 10.92YY18 pKa = 10.3KK19 pKa = 10.34GTSPNLAAFTRR30 pKa = 11.84HH31 pKa = 6.34AIHH34 pKa = 7.38LFFLWGKK41 pKa = 7.16TKK43 pKa = 9.66GWKK46 pKa = 6.47WTATAFKK53 pKa = 9.25LTRR56 pKa = 11.84LAITRR61 pKa = 11.84TLAGQEE67 pKa = 3.97LPRR70 pKa = 11.84PFGISLCKK78 pKa = 9.36KK79 pKa = 8.23TNLPMMLPLQVRR91 pKa = 11.84LAILRR96 pKa = 11.84KK97 pKa = 9.69DD98 pKa = 3.6FLLISYY104 pKa = 9.31VLTVLQLSRR113 pKa = 11.84LILGTGVIEE122 pKa = 4.74SYY124 pKa = 11.46DD125 pKa = 4.05SITRR129 pKa = 11.84PCTGSNPFTGASVSIVMKK147 pKa = 10.62RR148 pKa = 11.84MGIRR152 pKa = 11.84RR153 pKa = 11.84LDD155 pKa = 3.61EE156 pKa = 4.15PTSFTFPWVSTSGPNGISIATSFSDD181 pKa = 5.24LMNIPQPILEE191 pKa = 4.41CLYY194 pKa = 10.39TLGGPEE200 pKa = 4.17FKK202 pKa = 10.64EE203 pKa = 4.19KK204 pKa = 10.78VSSLKK209 pKa = 10.96GFVEE213 pKa = 4.61TTNGLLWKK221 pKa = 8.83LTKK224 pKa = 10.23LDD226 pKa = 3.8PDD228 pKa = 3.32KK229 pKa = 11.18TGNLLRR235 pKa = 11.84RR236 pKa = 11.84ISIKK240 pKa = 9.94KK241 pKa = 9.15DD242 pKa = 2.89KK243 pKa = 9.76EE244 pKa = 4.15YY245 pKa = 10.85KK246 pKa = 9.9SRR248 pKa = 11.84PFAIVDD254 pKa = 3.93YY255 pKa = 8.89ITQSALTPLHH265 pKa = 6.46DD266 pKa = 3.38RR267 pKa = 11.84LYY269 pKa = 10.35RR270 pKa = 11.84VLGSIPQDD278 pKa = 3.31CTFDD282 pKa = 3.49QNKK285 pKa = 9.3GFKK288 pKa = 10.18DD289 pKa = 3.49LLYY292 pKa = 11.07GGGPYY297 pKa = 10.63YY298 pKa = 11.11SFDD301 pKa = 3.51LTSATDD307 pKa = 3.65RR308 pKa = 11.84FPIFVQEE315 pKa = 4.98MVLAWLTSEE324 pKa = 4.48QYY326 pKa = 11.27ASAWVQAMVGIPFSTPNGPEE346 pKa = 3.78VEE348 pKa = 5.03FKK350 pKa = 10.81CGQPLGAKK358 pKa = 9.83SSWAMFTLSHH368 pKa = 6.42HH369 pKa = 6.58FVVQYY374 pKa = 10.63CAMVLNIDD382 pKa = 3.61NPRR385 pKa = 11.84YY386 pKa = 9.97KK387 pKa = 10.33ILGDD391 pKa = 4.46DD392 pKa = 4.66IVICDD397 pKa = 3.45HH398 pKa = 6.99ALAAKK403 pKa = 9.21YY404 pKa = 10.51LEE406 pKa = 4.38VMSQLGVEE414 pKa = 4.15ISSVKK419 pKa = 8.97THH421 pKa = 5.2VSEE424 pKa = 4.97NLFEE428 pKa = 4.05FAKK431 pKa = 10.57RR432 pKa = 11.84FGLQSVEE439 pKa = 3.69ISSFPITALVSDD451 pKa = 4.01INNYY455 pKa = 9.3VSIVATLATTAVEE468 pKa = 4.26RR469 pKa = 11.84GNLPLFVSGNTPRR482 pKa = 11.84FWEE485 pKa = 4.16SLLKK489 pKa = 10.78KK490 pKa = 9.49EE491 pKa = 4.81DD492 pKa = 3.4KK493 pKa = 10.37PRR495 pKa = 11.84LVNNLVHH502 pKa = 6.66SARR505 pKa = 11.84LLSYY509 pKa = 11.27LLMSNKK515 pKa = 9.6PYY517 pKa = 10.82LVAQSDD523 pKa = 4.08LLRR526 pKa = 11.84FAEE529 pKa = 4.21AAGFGGLAISQVFAAYY545 pKa = 9.2KK546 pKa = 10.15RR547 pKa = 11.84AVLYY551 pKa = 10.08MKK553 pKa = 10.52DD554 pKa = 3.55RR555 pKa = 11.84EE556 pKa = 4.08IDD558 pKa = 3.34KK559 pKa = 10.74LADD562 pKa = 3.0MSMRR566 pKa = 11.84FIRR569 pKa = 11.84PVQSMLSQIMFLPWRR584 pKa = 11.84GRR586 pKa = 11.84VTVDD590 pKa = 2.9YY591 pKa = 10.77RR592 pKa = 11.84KK593 pKa = 9.93FIPIIDD599 pKa = 4.58SIDD602 pKa = 3.83SNKK605 pKa = 10.13EE606 pKa = 3.46LYY608 pKa = 10.79LSFRR612 pKa = 11.84EE613 pKa = 4.19EE614 pKa = 3.87FEE616 pKa = 4.28KK617 pKa = 11.14ADD619 pKa = 5.3DD620 pKa = 4.09LVTLNKK626 pKa = 9.96VLDD629 pKa = 3.92DD630 pKa = 3.88HH631 pKa = 7.5PIRR634 pKa = 11.84PMPNLNGLQPNRR646 pKa = 11.84PKK648 pKa = 10.95LLVGQTRR655 pKa = 11.84AALIRR660 pKa = 11.84ALVKK664 pKa = 10.35EE665 pKa = 4.24LTMLAKK671 pKa = 10.72GEE673 pKa = 4.09EE674 pKa = 4.39PKK676 pKa = 11.0
MM1 pKa = 6.89TQPLVLKK8 pKa = 10.31LMKK11 pKa = 9.52WVRR14 pKa = 11.84KK15 pKa = 9.24YY16 pKa = 10.61YY17 pKa = 10.92YY18 pKa = 10.3KK19 pKa = 10.34GTSPNLAAFTRR30 pKa = 11.84HH31 pKa = 6.34AIHH34 pKa = 7.38LFFLWGKK41 pKa = 7.16TKK43 pKa = 9.66GWKK46 pKa = 6.47WTATAFKK53 pKa = 9.25LTRR56 pKa = 11.84LAITRR61 pKa = 11.84TLAGQEE67 pKa = 3.97LPRR70 pKa = 11.84PFGISLCKK78 pKa = 9.36KK79 pKa = 8.23TNLPMMLPLQVRR91 pKa = 11.84LAILRR96 pKa = 11.84KK97 pKa = 9.69DD98 pKa = 3.6FLLISYY104 pKa = 9.31VLTVLQLSRR113 pKa = 11.84LILGTGVIEE122 pKa = 4.74SYY124 pKa = 11.46DD125 pKa = 4.05SITRR129 pKa = 11.84PCTGSNPFTGASVSIVMKK147 pKa = 10.62RR148 pKa = 11.84MGIRR152 pKa = 11.84RR153 pKa = 11.84LDD155 pKa = 3.61EE156 pKa = 4.15PTSFTFPWVSTSGPNGISIATSFSDD181 pKa = 5.24LMNIPQPILEE191 pKa = 4.41CLYY194 pKa = 10.39TLGGPEE200 pKa = 4.17FKK202 pKa = 10.64EE203 pKa = 4.19KK204 pKa = 10.78VSSLKK209 pKa = 10.96GFVEE213 pKa = 4.61TTNGLLWKK221 pKa = 8.83LTKK224 pKa = 10.23LDD226 pKa = 3.8PDD228 pKa = 3.32KK229 pKa = 11.18TGNLLRR235 pKa = 11.84RR236 pKa = 11.84ISIKK240 pKa = 9.94KK241 pKa = 9.15DD242 pKa = 2.89KK243 pKa = 9.76EE244 pKa = 4.15YY245 pKa = 10.85KK246 pKa = 9.9SRR248 pKa = 11.84PFAIVDD254 pKa = 3.93YY255 pKa = 8.89ITQSALTPLHH265 pKa = 6.46DD266 pKa = 3.38RR267 pKa = 11.84LYY269 pKa = 10.35RR270 pKa = 11.84VLGSIPQDD278 pKa = 3.31CTFDD282 pKa = 3.49QNKK285 pKa = 9.3GFKK288 pKa = 10.18DD289 pKa = 3.49LLYY292 pKa = 11.07GGGPYY297 pKa = 10.63YY298 pKa = 11.11SFDD301 pKa = 3.51LTSATDD307 pKa = 3.65RR308 pKa = 11.84FPIFVQEE315 pKa = 4.98MVLAWLTSEE324 pKa = 4.48QYY326 pKa = 11.27ASAWVQAMVGIPFSTPNGPEE346 pKa = 3.78VEE348 pKa = 5.03FKK350 pKa = 10.81CGQPLGAKK358 pKa = 9.83SSWAMFTLSHH368 pKa = 6.42HH369 pKa = 6.58FVVQYY374 pKa = 10.63CAMVLNIDD382 pKa = 3.61NPRR385 pKa = 11.84YY386 pKa = 9.97KK387 pKa = 10.33ILGDD391 pKa = 4.46DD392 pKa = 4.66IVICDD397 pKa = 3.45HH398 pKa = 6.99ALAAKK403 pKa = 9.21YY404 pKa = 10.51LEE406 pKa = 4.38VMSQLGVEE414 pKa = 4.15ISSVKK419 pKa = 8.97THH421 pKa = 5.2VSEE424 pKa = 4.97NLFEE428 pKa = 4.05FAKK431 pKa = 10.57RR432 pKa = 11.84FGLQSVEE439 pKa = 3.69ISSFPITALVSDD451 pKa = 4.01INNYY455 pKa = 9.3VSIVATLATTAVEE468 pKa = 4.26RR469 pKa = 11.84GNLPLFVSGNTPRR482 pKa = 11.84FWEE485 pKa = 4.16SLLKK489 pKa = 10.78KK490 pKa = 9.49EE491 pKa = 4.81DD492 pKa = 3.4KK493 pKa = 10.37PRR495 pKa = 11.84LVNNLVHH502 pKa = 6.66SARR505 pKa = 11.84LLSYY509 pKa = 11.27LLMSNKK515 pKa = 9.6PYY517 pKa = 10.82LVAQSDD523 pKa = 4.08LLRR526 pKa = 11.84FAEE529 pKa = 4.21AAGFGGLAISQVFAAYY545 pKa = 9.2KK546 pKa = 10.15RR547 pKa = 11.84AVLYY551 pKa = 10.08MKK553 pKa = 10.52DD554 pKa = 3.55RR555 pKa = 11.84EE556 pKa = 4.08IDD558 pKa = 3.34KK559 pKa = 10.74LADD562 pKa = 3.0MSMRR566 pKa = 11.84FIRR569 pKa = 11.84PVQSMLSQIMFLPWRR584 pKa = 11.84GRR586 pKa = 11.84VTVDD590 pKa = 2.9YY591 pKa = 10.77RR592 pKa = 11.84KK593 pKa = 9.93FIPIIDD599 pKa = 4.58SIDD602 pKa = 3.83SNKK605 pKa = 10.13EE606 pKa = 3.46LYY608 pKa = 10.79LSFRR612 pKa = 11.84EE613 pKa = 4.19EE614 pKa = 3.87FEE616 pKa = 4.28KK617 pKa = 11.14ADD619 pKa = 5.3DD620 pKa = 4.09LVTLNKK626 pKa = 9.96VLDD629 pKa = 3.92DD630 pKa = 3.88HH631 pKa = 7.5PIRR634 pKa = 11.84PMPNLNGLQPNRR646 pKa = 11.84PKK648 pKa = 10.95LLVGQTRR655 pKa = 11.84AALIRR660 pKa = 11.84ALVKK664 pKa = 10.35EE665 pKa = 4.24LTMLAKK671 pKa = 10.72GEE673 pKa = 4.09EE674 pKa = 4.39PKK676 pKa = 11.0
Molecular weight: 76.31 kDa
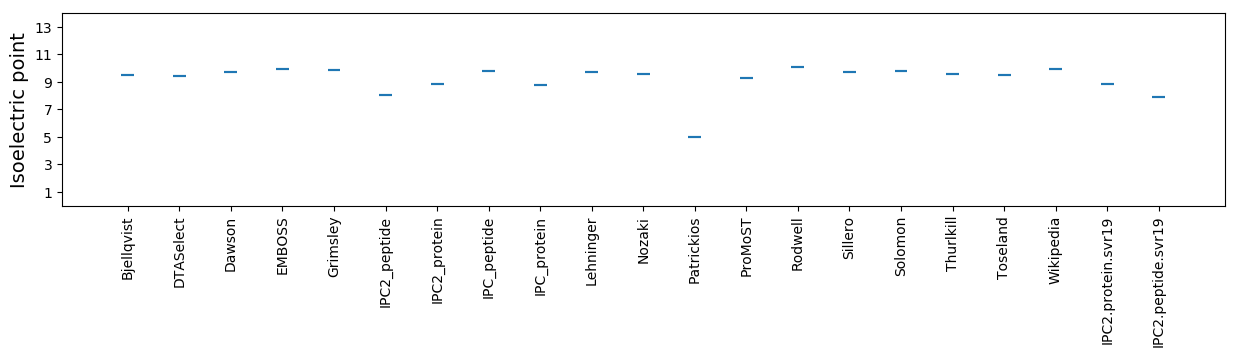
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H2EQQ4|H2EQQ4_9VIRU RNA-dependent RNA polymerases OS=Sclerotinia sclerotiorum mitovirus 2 OX=1133728 GN=SsMV2_gp1 PE=4 SV=1
MM1 pKa = 6.89TQPLVLKK8 pKa = 10.31LMKK11 pKa = 9.52WVRR14 pKa = 11.84KK15 pKa = 9.24YY16 pKa = 10.61YY17 pKa = 10.92YY18 pKa = 10.3KK19 pKa = 10.34GTSPNLAAFTRR30 pKa = 11.84HH31 pKa = 6.34AIHH34 pKa = 7.38LFFLWGKK41 pKa = 7.16TKK43 pKa = 9.66GWKK46 pKa = 6.47WTATAFKK53 pKa = 9.25LTRR56 pKa = 11.84LAITRR61 pKa = 11.84TLAGQEE67 pKa = 3.97LPRR70 pKa = 11.84PFGISLCKK78 pKa = 9.36KK79 pKa = 8.23TNLPMMLPLQVRR91 pKa = 11.84LAILRR96 pKa = 11.84KK97 pKa = 9.69DD98 pKa = 3.6FLLISYY104 pKa = 9.31VLTVLQLSRR113 pKa = 11.84LILGTGVIEE122 pKa = 4.74SYY124 pKa = 11.46DD125 pKa = 4.05SITRR129 pKa = 11.84PCTGSNPFTGASVSIVMKK147 pKa = 10.62RR148 pKa = 11.84MGIRR152 pKa = 11.84RR153 pKa = 11.84LDD155 pKa = 3.61EE156 pKa = 4.15PTSFTFPWVSTSGPNGISIATSFSDD181 pKa = 5.24LMNIPQPILEE191 pKa = 4.41CLYY194 pKa = 10.39TLGGPEE200 pKa = 4.17FKK202 pKa = 10.64EE203 pKa = 4.19KK204 pKa = 10.78VSSLKK209 pKa = 10.96GFVEE213 pKa = 4.61TTNGLLWKK221 pKa = 8.83LTKK224 pKa = 10.23LDD226 pKa = 3.8PDD228 pKa = 3.32KK229 pKa = 11.18TGNLLRR235 pKa = 11.84RR236 pKa = 11.84ISIKK240 pKa = 9.94KK241 pKa = 9.15DD242 pKa = 2.89KK243 pKa = 9.76EE244 pKa = 4.15YY245 pKa = 10.85KK246 pKa = 9.9SRR248 pKa = 11.84PFAIVDD254 pKa = 3.93YY255 pKa = 8.89ITQSALTPLHH265 pKa = 6.46DD266 pKa = 3.38RR267 pKa = 11.84LYY269 pKa = 10.35RR270 pKa = 11.84VLGSIPQDD278 pKa = 3.31CTFDD282 pKa = 3.49QNKK285 pKa = 9.3GFKK288 pKa = 10.18DD289 pKa = 3.49LLYY292 pKa = 11.07GGGPYY297 pKa = 10.63YY298 pKa = 11.11SFDD301 pKa = 3.51LTSATDD307 pKa = 3.65RR308 pKa = 11.84FPIFVQEE315 pKa = 4.98MVLAWLTSEE324 pKa = 4.48QYY326 pKa = 11.27ASAWVQAMVGIPFSTPNGPEE346 pKa = 3.78VEE348 pKa = 5.03FKK350 pKa = 10.81CGQPLGAKK358 pKa = 9.83SSWAMFTLSHH368 pKa = 6.42HH369 pKa = 6.58FVVQYY374 pKa = 10.63CAMVLNIDD382 pKa = 3.61NPRR385 pKa = 11.84YY386 pKa = 9.97KK387 pKa = 10.33ILGDD391 pKa = 4.46DD392 pKa = 4.66IVICDD397 pKa = 3.45HH398 pKa = 6.99ALAAKK403 pKa = 9.21YY404 pKa = 10.51LEE406 pKa = 4.38VMSQLGVEE414 pKa = 4.15ISSVKK419 pKa = 8.97THH421 pKa = 5.2VSEE424 pKa = 4.97NLFEE428 pKa = 4.05FAKK431 pKa = 10.57RR432 pKa = 11.84FGLQSVEE439 pKa = 3.69ISSFPITALVSDD451 pKa = 4.01INNYY455 pKa = 9.3VSIVATLATTAVEE468 pKa = 4.26RR469 pKa = 11.84GNLPLFVSGNTPRR482 pKa = 11.84FWEE485 pKa = 4.16SLLKK489 pKa = 10.78KK490 pKa = 9.49EE491 pKa = 4.81DD492 pKa = 3.4KK493 pKa = 10.37PRR495 pKa = 11.84LVNNLVHH502 pKa = 6.66SARR505 pKa = 11.84LLSYY509 pKa = 11.27LLMSNKK515 pKa = 9.6PYY517 pKa = 10.82LVAQSDD523 pKa = 4.08LLRR526 pKa = 11.84FAEE529 pKa = 4.21AAGFGGLAISQVFAAYY545 pKa = 9.2KK546 pKa = 10.15RR547 pKa = 11.84AVLYY551 pKa = 10.08MKK553 pKa = 10.52DD554 pKa = 3.55RR555 pKa = 11.84EE556 pKa = 4.08IDD558 pKa = 3.34KK559 pKa = 10.74LADD562 pKa = 3.0MSMRR566 pKa = 11.84FIRR569 pKa = 11.84PVQSMLSQIMFLPWRR584 pKa = 11.84GRR586 pKa = 11.84VTVDD590 pKa = 2.9YY591 pKa = 10.77RR592 pKa = 11.84KK593 pKa = 9.93FIPIIDD599 pKa = 4.58SIDD602 pKa = 3.83SNKK605 pKa = 10.13EE606 pKa = 3.46LYY608 pKa = 10.79LSFRR612 pKa = 11.84EE613 pKa = 4.19EE614 pKa = 3.87FEE616 pKa = 4.28KK617 pKa = 11.14ADD619 pKa = 5.3DD620 pKa = 4.09LVTLNKK626 pKa = 9.96VLDD629 pKa = 3.92DD630 pKa = 3.88HH631 pKa = 7.5PIRR634 pKa = 11.84PMPNLNGLQPNRR646 pKa = 11.84PKK648 pKa = 10.95LLVGQTRR655 pKa = 11.84AALIRR660 pKa = 11.84ALVKK664 pKa = 10.35EE665 pKa = 4.24LTMLAKK671 pKa = 10.72GEE673 pKa = 4.09EE674 pKa = 4.39PKK676 pKa = 11.0
MM1 pKa = 6.89TQPLVLKK8 pKa = 10.31LMKK11 pKa = 9.52WVRR14 pKa = 11.84KK15 pKa = 9.24YY16 pKa = 10.61YY17 pKa = 10.92YY18 pKa = 10.3KK19 pKa = 10.34GTSPNLAAFTRR30 pKa = 11.84HH31 pKa = 6.34AIHH34 pKa = 7.38LFFLWGKK41 pKa = 7.16TKK43 pKa = 9.66GWKK46 pKa = 6.47WTATAFKK53 pKa = 9.25LTRR56 pKa = 11.84LAITRR61 pKa = 11.84TLAGQEE67 pKa = 3.97LPRR70 pKa = 11.84PFGISLCKK78 pKa = 9.36KK79 pKa = 8.23TNLPMMLPLQVRR91 pKa = 11.84LAILRR96 pKa = 11.84KK97 pKa = 9.69DD98 pKa = 3.6FLLISYY104 pKa = 9.31VLTVLQLSRR113 pKa = 11.84LILGTGVIEE122 pKa = 4.74SYY124 pKa = 11.46DD125 pKa = 4.05SITRR129 pKa = 11.84PCTGSNPFTGASVSIVMKK147 pKa = 10.62RR148 pKa = 11.84MGIRR152 pKa = 11.84RR153 pKa = 11.84LDD155 pKa = 3.61EE156 pKa = 4.15PTSFTFPWVSTSGPNGISIATSFSDD181 pKa = 5.24LMNIPQPILEE191 pKa = 4.41CLYY194 pKa = 10.39TLGGPEE200 pKa = 4.17FKK202 pKa = 10.64EE203 pKa = 4.19KK204 pKa = 10.78VSSLKK209 pKa = 10.96GFVEE213 pKa = 4.61TTNGLLWKK221 pKa = 8.83LTKK224 pKa = 10.23LDD226 pKa = 3.8PDD228 pKa = 3.32KK229 pKa = 11.18TGNLLRR235 pKa = 11.84RR236 pKa = 11.84ISIKK240 pKa = 9.94KK241 pKa = 9.15DD242 pKa = 2.89KK243 pKa = 9.76EE244 pKa = 4.15YY245 pKa = 10.85KK246 pKa = 9.9SRR248 pKa = 11.84PFAIVDD254 pKa = 3.93YY255 pKa = 8.89ITQSALTPLHH265 pKa = 6.46DD266 pKa = 3.38RR267 pKa = 11.84LYY269 pKa = 10.35RR270 pKa = 11.84VLGSIPQDD278 pKa = 3.31CTFDD282 pKa = 3.49QNKK285 pKa = 9.3GFKK288 pKa = 10.18DD289 pKa = 3.49LLYY292 pKa = 11.07GGGPYY297 pKa = 10.63YY298 pKa = 11.11SFDD301 pKa = 3.51LTSATDD307 pKa = 3.65RR308 pKa = 11.84FPIFVQEE315 pKa = 4.98MVLAWLTSEE324 pKa = 4.48QYY326 pKa = 11.27ASAWVQAMVGIPFSTPNGPEE346 pKa = 3.78VEE348 pKa = 5.03FKK350 pKa = 10.81CGQPLGAKK358 pKa = 9.83SSWAMFTLSHH368 pKa = 6.42HH369 pKa = 6.58FVVQYY374 pKa = 10.63CAMVLNIDD382 pKa = 3.61NPRR385 pKa = 11.84YY386 pKa = 9.97KK387 pKa = 10.33ILGDD391 pKa = 4.46DD392 pKa = 4.66IVICDD397 pKa = 3.45HH398 pKa = 6.99ALAAKK403 pKa = 9.21YY404 pKa = 10.51LEE406 pKa = 4.38VMSQLGVEE414 pKa = 4.15ISSVKK419 pKa = 8.97THH421 pKa = 5.2VSEE424 pKa = 4.97NLFEE428 pKa = 4.05FAKK431 pKa = 10.57RR432 pKa = 11.84FGLQSVEE439 pKa = 3.69ISSFPITALVSDD451 pKa = 4.01INNYY455 pKa = 9.3VSIVATLATTAVEE468 pKa = 4.26RR469 pKa = 11.84GNLPLFVSGNTPRR482 pKa = 11.84FWEE485 pKa = 4.16SLLKK489 pKa = 10.78KK490 pKa = 9.49EE491 pKa = 4.81DD492 pKa = 3.4KK493 pKa = 10.37PRR495 pKa = 11.84LVNNLVHH502 pKa = 6.66SARR505 pKa = 11.84LLSYY509 pKa = 11.27LLMSNKK515 pKa = 9.6PYY517 pKa = 10.82LVAQSDD523 pKa = 4.08LLRR526 pKa = 11.84FAEE529 pKa = 4.21AAGFGGLAISQVFAAYY545 pKa = 9.2KK546 pKa = 10.15RR547 pKa = 11.84AVLYY551 pKa = 10.08MKK553 pKa = 10.52DD554 pKa = 3.55RR555 pKa = 11.84EE556 pKa = 4.08IDD558 pKa = 3.34KK559 pKa = 10.74LADD562 pKa = 3.0MSMRR566 pKa = 11.84FIRR569 pKa = 11.84PVQSMLSQIMFLPWRR584 pKa = 11.84GRR586 pKa = 11.84VTVDD590 pKa = 2.9YY591 pKa = 10.77RR592 pKa = 11.84KK593 pKa = 9.93FIPIIDD599 pKa = 4.58SIDD602 pKa = 3.83SNKK605 pKa = 10.13EE606 pKa = 3.46LYY608 pKa = 10.79LSFRR612 pKa = 11.84EE613 pKa = 4.19EE614 pKa = 3.87FEE616 pKa = 4.28KK617 pKa = 11.14ADD619 pKa = 5.3DD620 pKa = 4.09LVTLNKK626 pKa = 9.96VLDD629 pKa = 3.92DD630 pKa = 3.88HH631 pKa = 7.5PIRR634 pKa = 11.84PMPNLNGLQPNRR646 pKa = 11.84PKK648 pKa = 10.95LLVGQTRR655 pKa = 11.84AALIRR660 pKa = 11.84ALVKK664 pKa = 10.35EE665 pKa = 4.24LTMLAKK671 pKa = 10.72GEE673 pKa = 4.09EE674 pKa = 4.39PKK676 pKa = 11.0
Molecular weight: 76.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
676 |
676 |
676 |
676.0 |
76.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.361 ± 0.0 | 1.036 ± 0.0 |
4.586 ± 0.0 | 4.29 ± 0.0 |
5.325 ± 0.0 | 5.621 ± 0.0 |
1.331 ± 0.0 | 5.917 ± 0.0 |
6.657 ± 0.0 | 12.722 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.959 ± 0.0 | 3.55 ± 0.0 |
5.769 ± 0.0 | 3.107 ± 0.0 |
5.473 ± 0.0 | 7.544 ± 0.0 |
6.213 ± 0.0 | 6.509 ± 0.0 |
1.627 ± 0.0 | 3.402 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
