
Georgenia subflava
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Bogoriellaceae; Georgenia
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
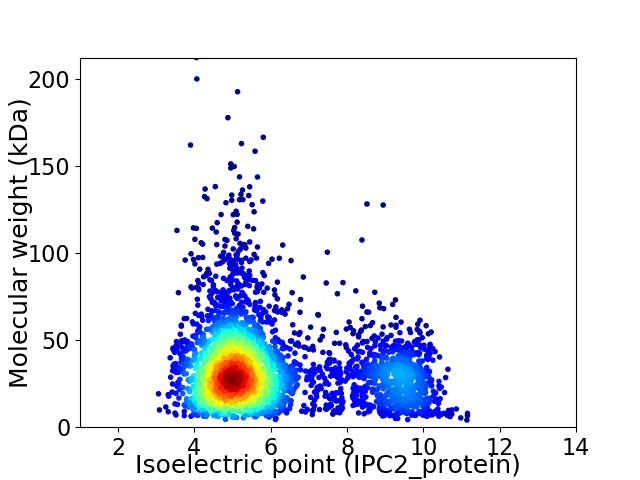
Virtual 2D-PAGE plot for 3778 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N7EHI8|A0A6N7EHI8_9MICO HPr family phosphocarrier protein OS=Georgenia subflava OX=1622177 GN=GB881_12455 PE=4 SV=1
MM1 pKa = 7.51APAGKK6 pKa = 9.57SRR8 pKa = 11.84RR9 pKa = 11.84AMVFGATAAFALVLAGCAEE28 pKa = 4.24TEE30 pKa = 4.18DD31 pKa = 4.35PDD33 pKa = 5.02GGGDD37 pKa = 3.42ATGGTDD43 pKa = 3.07GGSSEE48 pKa = 4.89GITVGTTDD56 pKa = 4.52VITNLDD62 pKa = 3.61PAGSYY67 pKa = 11.31DD68 pKa = 3.46NGSFAVQNQVFPFIMNTPYY87 pKa = 10.86GSPDD91 pKa = 3.56VEE93 pKa = 3.89PDD95 pKa = 3.01IAEE98 pKa = 4.28SAEE101 pKa = 4.0FTAPTEE107 pKa = 4.05YY108 pKa = 9.96TVKK111 pKa = 10.56LKK113 pKa = 10.97EE114 pKa = 4.14GLTFANGNEE123 pKa = 4.04LTASDD128 pKa = 3.97VKK130 pKa = 10.5FTFDD134 pKa = 3.24RR135 pKa = 11.84QIAIADD141 pKa = 3.66PSGPSSLLYY150 pKa = 10.79NLDD153 pKa = 3.48SVEE156 pKa = 4.25VVDD159 pKa = 5.06DD160 pKa = 3.82LTVVFHH166 pKa = 6.82LKK168 pKa = 10.4SEE170 pKa = 4.12NDD172 pKa = 3.26QVFPQILSSPAGPIVDD188 pKa = 3.91EE189 pKa = 5.9DD190 pKa = 4.14VFLPDD195 pKa = 4.21AVTPADD201 pKa = 5.13DD202 pKa = 3.78IVSGNAFAGQYY213 pKa = 10.81AITDD217 pKa = 3.58YY218 pKa = 11.27TEE220 pKa = 4.27NEE222 pKa = 4.39LIQYY226 pKa = 8.18QAFDD230 pKa = 4.31GYY232 pKa = 11.39DD233 pKa = 3.38GVLGAARR240 pKa = 11.84TDD242 pKa = 3.5TVTAQYY248 pKa = 10.32YY249 pKa = 10.56AEE251 pKa = 4.09EE252 pKa = 4.38TSLKK256 pKa = 10.64LAVQEE261 pKa = 4.16GDD263 pKa = 2.62IDD265 pKa = 3.88VAFRR269 pKa = 11.84SLSPTDD275 pKa = 3.79LADD278 pKa = 3.57LRR280 pKa = 11.84EE281 pKa = 4.47DD282 pKa = 3.82EE283 pKa = 4.77NVTVHH288 pKa = 7.11DD289 pKa = 4.85GPGGEE294 pKa = 3.61IRR296 pKa = 11.84YY297 pKa = 9.33IVFNFNTQPFGAEE310 pKa = 3.58TVEE313 pKa = 3.98ADD315 pKa = 3.43EE316 pKa = 4.89AKK318 pKa = 10.78ALAVRR323 pKa = 11.84QAVAHH328 pKa = 5.86SLDD331 pKa = 3.7RR332 pKa = 11.84DD333 pKa = 3.63ALSNEE338 pKa = 4.07IYY340 pKa = 10.54NGSFTPLYY348 pKa = 10.3SYY350 pKa = 11.07VPEE353 pKa = 4.54GLTGAVEE360 pKa = 4.14PLKK363 pKa = 10.6EE364 pKa = 4.24LYY366 pKa = 10.67GDD368 pKa = 3.93GAGGPDD374 pKa = 3.76ADD376 pKa = 3.77AAAAVLEE383 pKa = 4.37EE384 pKa = 4.87AGVEE388 pKa = 4.27TPVALNLQFSPDD400 pKa = 3.3HH401 pKa = 6.4YY402 pKa = 11.27GNSSDD407 pKa = 5.57LEE409 pKa = 4.14YY410 pKa = 11.46ALIEE414 pKa = 4.37SQLEE418 pKa = 3.88AGGLFDD424 pKa = 3.72VTLNSSLWDD433 pKa = 3.66AYY435 pKa = 9.75ATEE438 pKa = 4.71RR439 pKa = 11.84LTDD442 pKa = 3.59YY443 pKa = 10.91PSYY446 pKa = 10.34QLGWFPDD453 pKa = 3.67YY454 pKa = 11.48SDD456 pKa = 4.11ADD458 pKa = 3.57NYY460 pKa = 8.94LTPFFLIEE468 pKa = 3.99NFLGNGYY475 pKa = 10.87ANQEE479 pKa = 4.15VNDD482 pKa = 5.25LILDD486 pKa = 3.75QATTADD492 pKa = 3.68PDD494 pKa = 3.34EE495 pKa = 4.52RR496 pKa = 11.84AGKK499 pKa = 9.79IEE501 pKa = 4.0QIQEE505 pKa = 3.92LVAQDD510 pKa = 4.58LSTVPYY516 pKa = 10.3LQGAQVAVAATDD528 pKa = 3.35ITGVTDD534 pKa = 3.38TLDD537 pKa = 3.32ASFKK541 pKa = 10.4FRR543 pKa = 11.84YY544 pKa = 9.01AALGRR549 pKa = 3.94
MM1 pKa = 7.51APAGKK6 pKa = 9.57SRR8 pKa = 11.84RR9 pKa = 11.84AMVFGATAAFALVLAGCAEE28 pKa = 4.24TEE30 pKa = 4.18DD31 pKa = 4.35PDD33 pKa = 5.02GGGDD37 pKa = 3.42ATGGTDD43 pKa = 3.07GGSSEE48 pKa = 4.89GITVGTTDD56 pKa = 4.52VITNLDD62 pKa = 3.61PAGSYY67 pKa = 11.31DD68 pKa = 3.46NGSFAVQNQVFPFIMNTPYY87 pKa = 10.86GSPDD91 pKa = 3.56VEE93 pKa = 3.89PDD95 pKa = 3.01IAEE98 pKa = 4.28SAEE101 pKa = 4.0FTAPTEE107 pKa = 4.05YY108 pKa = 9.96TVKK111 pKa = 10.56LKK113 pKa = 10.97EE114 pKa = 4.14GLTFANGNEE123 pKa = 4.04LTASDD128 pKa = 3.97VKK130 pKa = 10.5FTFDD134 pKa = 3.24RR135 pKa = 11.84QIAIADD141 pKa = 3.66PSGPSSLLYY150 pKa = 10.79NLDD153 pKa = 3.48SVEE156 pKa = 4.25VVDD159 pKa = 5.06DD160 pKa = 3.82LTVVFHH166 pKa = 6.82LKK168 pKa = 10.4SEE170 pKa = 4.12NDD172 pKa = 3.26QVFPQILSSPAGPIVDD188 pKa = 3.91EE189 pKa = 5.9DD190 pKa = 4.14VFLPDD195 pKa = 4.21AVTPADD201 pKa = 5.13DD202 pKa = 3.78IVSGNAFAGQYY213 pKa = 10.81AITDD217 pKa = 3.58YY218 pKa = 11.27TEE220 pKa = 4.27NEE222 pKa = 4.39LIQYY226 pKa = 8.18QAFDD230 pKa = 4.31GYY232 pKa = 11.39DD233 pKa = 3.38GVLGAARR240 pKa = 11.84TDD242 pKa = 3.5TVTAQYY248 pKa = 10.32YY249 pKa = 10.56AEE251 pKa = 4.09EE252 pKa = 4.38TSLKK256 pKa = 10.64LAVQEE261 pKa = 4.16GDD263 pKa = 2.62IDD265 pKa = 3.88VAFRR269 pKa = 11.84SLSPTDD275 pKa = 3.79LADD278 pKa = 3.57LRR280 pKa = 11.84EE281 pKa = 4.47DD282 pKa = 3.82EE283 pKa = 4.77NVTVHH288 pKa = 7.11DD289 pKa = 4.85GPGGEE294 pKa = 3.61IRR296 pKa = 11.84YY297 pKa = 9.33IVFNFNTQPFGAEE310 pKa = 3.58TVEE313 pKa = 3.98ADD315 pKa = 3.43EE316 pKa = 4.89AKK318 pKa = 10.78ALAVRR323 pKa = 11.84QAVAHH328 pKa = 5.86SLDD331 pKa = 3.7RR332 pKa = 11.84DD333 pKa = 3.63ALSNEE338 pKa = 4.07IYY340 pKa = 10.54NGSFTPLYY348 pKa = 10.3SYY350 pKa = 11.07VPEE353 pKa = 4.54GLTGAVEE360 pKa = 4.14PLKK363 pKa = 10.6EE364 pKa = 4.24LYY366 pKa = 10.67GDD368 pKa = 3.93GAGGPDD374 pKa = 3.76ADD376 pKa = 3.77AAAAVLEE383 pKa = 4.37EE384 pKa = 4.87AGVEE388 pKa = 4.27TPVALNLQFSPDD400 pKa = 3.3HH401 pKa = 6.4YY402 pKa = 11.27GNSSDD407 pKa = 5.57LEE409 pKa = 4.14YY410 pKa = 11.46ALIEE414 pKa = 4.37SQLEE418 pKa = 3.88AGGLFDD424 pKa = 3.72VTLNSSLWDD433 pKa = 3.66AYY435 pKa = 9.75ATEE438 pKa = 4.71RR439 pKa = 11.84LTDD442 pKa = 3.59YY443 pKa = 10.91PSYY446 pKa = 10.34QLGWFPDD453 pKa = 3.67YY454 pKa = 11.48SDD456 pKa = 4.11ADD458 pKa = 3.57NYY460 pKa = 8.94LTPFFLIEE468 pKa = 3.99NFLGNGYY475 pKa = 10.87ANQEE479 pKa = 4.15VNDD482 pKa = 5.25LILDD486 pKa = 3.75QATTADD492 pKa = 3.68PDD494 pKa = 3.34EE495 pKa = 4.52RR496 pKa = 11.84AGKK499 pKa = 9.79IEE501 pKa = 4.0QIQEE505 pKa = 3.92LVAQDD510 pKa = 4.58LSTVPYY516 pKa = 10.3LQGAQVAVAATDD528 pKa = 3.35ITGVTDD534 pKa = 3.38TLDD537 pKa = 3.32ASFKK541 pKa = 10.4FRR543 pKa = 11.84YY544 pKa = 9.01AALGRR549 pKa = 3.94
Molecular weight: 58.53 kDa
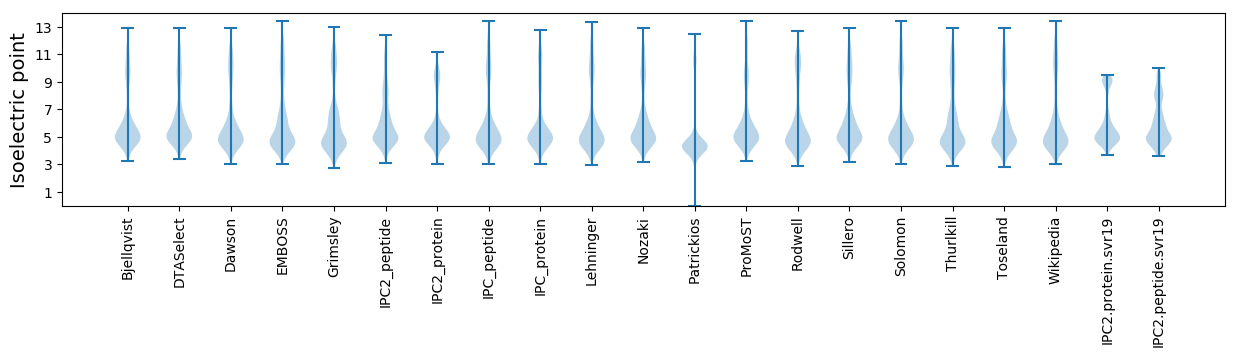
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N7EQX2|A0A6N7EQX2_9MICO Alpha/beta fold hydrolase OS=Georgenia subflava OX=1622177 GN=GB881_18100 PE=3 SV=1
NN1 pKa = 7.57RR2 pKa = 11.84PPRR5 pKa = 11.84PRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84GRR11 pKa = 11.84GRR13 pKa = 11.84RR14 pKa = 11.84PRR16 pKa = 11.84GARR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84PARR25 pKa = 11.84RR26 pKa = 11.84SPAGAAPTSSRR37 pKa = 11.84PHH39 pKa = 6.57APRR42 pKa = 11.84PLTSRR47 pKa = 11.84SGTVAEE53 pKa = 4.53PRR55 pKa = 11.84LTARR59 pKa = 11.84GG60 pKa = 3.52
NN1 pKa = 7.57RR2 pKa = 11.84PPRR5 pKa = 11.84PRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84GRR11 pKa = 11.84GRR13 pKa = 11.84RR14 pKa = 11.84PRR16 pKa = 11.84GARR19 pKa = 11.84PRR21 pKa = 11.84RR22 pKa = 11.84PARR25 pKa = 11.84RR26 pKa = 11.84SPAGAAPTSSRR37 pKa = 11.84PHH39 pKa = 6.57APRR42 pKa = 11.84PLTSRR47 pKa = 11.84SGTVAEE53 pKa = 4.53PRR55 pKa = 11.84LTARR59 pKa = 11.84GG60 pKa = 3.52
Molecular weight: 6.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1231966 |
32 |
2007 |
326.1 |
34.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.927 ± 0.057 | 0.532 ± 0.008 |
6.357 ± 0.032 | 5.91 ± 0.038 |
2.567 ± 0.022 | 9.475 ± 0.041 |
2.176 ± 0.019 | 3.227 ± 0.025 |
1.485 ± 0.025 | 10.326 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.798 ± 0.015 | 1.639 ± 0.019 |
5.749 ± 0.036 | 2.65 ± 0.021 |
7.769 ± 0.044 | 4.967 ± 0.025 |
6.368 ± 0.031 | 9.73 ± 0.042 |
1.47 ± 0.018 | 1.879 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
