
Staphylococcus phage BP39
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Rountreeviridae; Rakietenvirinae; Rosenblumvirus; Staphylococcus virus BP39
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
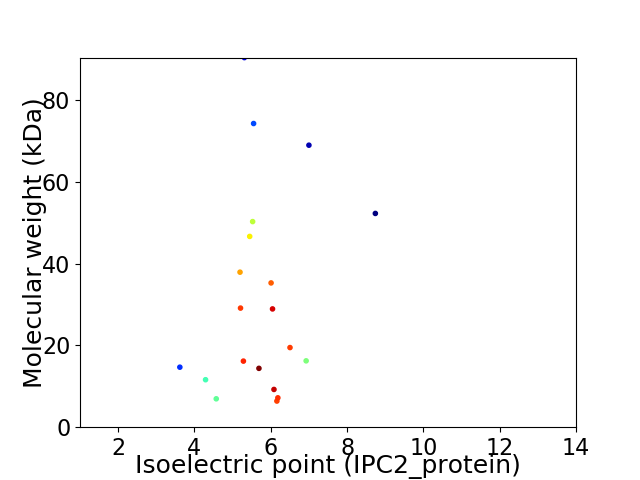
Virtual 2D-PAGE plot for 20 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A185AMV8|A0A185AMV8_9CAUD Uncharacterized protein OS=Staphylococcus phage BP39 OX=1543206 GN=BP39_03 PE=4 SV=1
MM1 pKa = 7.27VNVDD5 pKa = 3.72NAPEE9 pKa = 4.15EE10 pKa = 4.21KK11 pKa = 10.3GQAYY15 pKa = 8.38TEE17 pKa = 3.92MLQLFNKK24 pKa = 9.67LIQWNPAYY32 pKa = 9.81TFDD35 pKa = 3.95NAINLVSACQQLLLNYY51 pKa = 9.28NSSVVKK57 pKa = 9.87FLNDD61 pKa = 3.22EE62 pKa = 4.62LNNEE66 pKa = 4.41TKK68 pKa = 10.41PEE70 pKa = 4.48SILSYY75 pKa = 10.51IAGDD79 pKa = 4.14DD80 pKa = 5.49SIEE83 pKa = 3.7QWNMHH88 pKa = 5.76KK89 pKa = 10.66GFYY92 pKa = 7.63EE93 pKa = 4.27TYY95 pKa = 9.67NVYY98 pKa = 10.34VFF100 pKa = 4.47
MM1 pKa = 7.27VNVDD5 pKa = 3.72NAPEE9 pKa = 4.15EE10 pKa = 4.21KK11 pKa = 10.3GQAYY15 pKa = 8.38TEE17 pKa = 3.92MLQLFNKK24 pKa = 9.67LIQWNPAYY32 pKa = 9.81TFDD35 pKa = 3.95NAINLVSACQQLLLNYY51 pKa = 9.28NSSVVKK57 pKa = 9.87FLNDD61 pKa = 3.22EE62 pKa = 4.62LNNEE66 pKa = 4.41TKK68 pKa = 10.41PEE70 pKa = 4.48SILSYY75 pKa = 10.51IAGDD79 pKa = 4.14DD80 pKa = 5.49SIEE83 pKa = 3.7QWNMHH88 pKa = 5.76KK89 pKa = 10.66GFYY92 pKa = 7.63EE93 pKa = 4.27TYY95 pKa = 9.67NVYY98 pKa = 10.34VFF100 pKa = 4.47
Molecular weight: 11.61 kDa
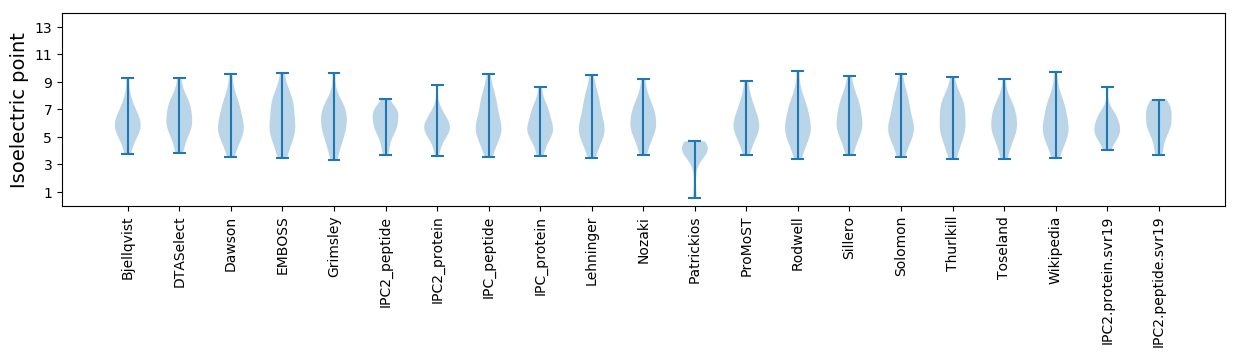
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A185AMW6|A0A185AMW6_9CAUD Holin OS=Staphylococcus phage BP39 OX=1543206 GN=BP39_11 PE=4 SV=1
MM1 pKa = 7.66NDD3 pKa = 2.99QEE5 pKa = 5.77KK6 pKa = 10.06IDD8 pKa = 4.58KK9 pKa = 7.93FTHH12 pKa = 6.46SYY14 pKa = 10.93INDD17 pKa = 3.77DD18 pKa = 3.78FGLTIDD24 pKa = 3.66QLVPKK29 pKa = 9.85VKK31 pKa = 10.36GYY33 pKa = 11.2GRR35 pKa = 11.84FNVWLGGNEE44 pKa = 4.07SKK46 pKa = 10.39IRR48 pKa = 11.84QVLKK52 pKa = 9.89AVKK55 pKa = 9.61EE56 pKa = 4.15IGVSPTLFAVYY67 pKa = 9.33EE68 pKa = 4.19KK69 pKa = 11.33NEE71 pKa = 4.39GYY73 pKa = 10.08SAGLGWLNHH82 pKa = 4.64TSARR86 pKa = 11.84GDD88 pKa = 3.67YY89 pKa = 9.83LTDD92 pKa = 3.0AKK94 pKa = 10.81FIARR98 pKa = 11.84KK99 pKa = 9.11LVSQSKK105 pKa = 9.27QAGQPSWYY113 pKa = 9.92DD114 pKa = 3.2YY115 pKa = 11.1GNPVHH120 pKa = 7.0FVPQDD125 pKa = 3.43VQRR128 pKa = 11.84KK129 pKa = 8.58GNADD133 pKa = 3.15FSKK136 pKa = 10.88NMKK139 pKa = 10.18AGTIGRR145 pKa = 11.84AYY147 pKa = 9.86IPLTAAATWAAYY159 pKa = 10.19YY160 pKa = 9.97PLGLKK165 pKa = 10.27ASYY168 pKa = 11.1NKK170 pKa = 8.87VQNYY174 pKa = 8.36GNPFLDD180 pKa = 4.01GANTILAWGGKK191 pKa = 8.9IDD193 pKa = 4.74GKK195 pKa = 10.89GGSPNSGSSDD205 pKa = 3.32SSSDD209 pKa = 3.31SGGNSLLALAKK220 pKa = 10.19QAMQEE225 pKa = 4.1LLKK228 pKa = 10.63KK229 pKa = 10.48VQDD232 pKa = 3.53ALQWDD237 pKa = 4.06VHH239 pKa = 7.39SIGSDD244 pKa = 2.68KK245 pKa = 10.79FFSNDD250 pKa = 2.89YY251 pKa = 8.37FTLQKK256 pKa = 9.76TFNNTYY262 pKa = 9.12HH263 pKa = 7.0IKK265 pKa = 8.04MTIGLLDD272 pKa = 3.83SLKK275 pKa = 10.93KK276 pKa = 10.89LIDD279 pKa = 3.73SVQVDD284 pKa = 3.66SGGSSSNPTDD294 pKa = 4.41DD295 pKa = 6.01DD296 pKa = 4.66GDD298 pKa = 3.93HH299 pKa = 6.69KK300 pKa = 10.7PISGKK305 pKa = 7.43SVKK308 pKa = 10.22PNGKK312 pKa = 8.68SGRR315 pKa = 11.84VIGGNWTYY323 pKa = 11.51AQLPEE328 pKa = 4.51KK329 pKa = 9.49YY330 pKa = 9.06KK331 pKa = 10.77KK332 pKa = 10.74AIGVPLFKK340 pKa = 10.35KK341 pKa = 10.27EE342 pKa = 3.9YY343 pKa = 9.69LYY345 pKa = 11.13KK346 pKa = 10.3PGNIFPQTGNAGQCTEE362 pKa = 4.3LTWAYY367 pKa = 9.46MSQLHH372 pKa = 6.84GKK374 pKa = 8.57RR375 pKa = 11.84QPTDD379 pKa = 3.37DD380 pKa = 3.88GQVTNGQRR388 pKa = 11.84VWYY391 pKa = 9.59VYY393 pKa = 10.68KK394 pKa = 10.88KK395 pKa = 10.51LGAKK399 pKa = 6.42TTHH402 pKa = 6.34NPTVGYY408 pKa = 9.76GFSSKK413 pKa = 10.35PPYY416 pKa = 10.08LQATAYY422 pKa = 10.12GIGHH426 pKa = 6.67TGVVVAVFDD435 pKa = 4.62DD436 pKa = 4.68GSFLVANYY444 pKa = 8.94NVPPYY449 pKa = 9.33VAPSRR454 pKa = 11.84VVLYY458 pKa = 8.72TLINGVPHH466 pKa = 6.57NAGDD470 pKa = 3.9NIVFFSGIAA479 pKa = 3.09
MM1 pKa = 7.66NDD3 pKa = 2.99QEE5 pKa = 5.77KK6 pKa = 10.06IDD8 pKa = 4.58KK9 pKa = 7.93FTHH12 pKa = 6.46SYY14 pKa = 10.93INDD17 pKa = 3.77DD18 pKa = 3.78FGLTIDD24 pKa = 3.66QLVPKK29 pKa = 9.85VKK31 pKa = 10.36GYY33 pKa = 11.2GRR35 pKa = 11.84FNVWLGGNEE44 pKa = 4.07SKK46 pKa = 10.39IRR48 pKa = 11.84QVLKK52 pKa = 9.89AVKK55 pKa = 9.61EE56 pKa = 4.15IGVSPTLFAVYY67 pKa = 9.33EE68 pKa = 4.19KK69 pKa = 11.33NEE71 pKa = 4.39GYY73 pKa = 10.08SAGLGWLNHH82 pKa = 4.64TSARR86 pKa = 11.84GDD88 pKa = 3.67YY89 pKa = 9.83LTDD92 pKa = 3.0AKK94 pKa = 10.81FIARR98 pKa = 11.84KK99 pKa = 9.11LVSQSKK105 pKa = 9.27QAGQPSWYY113 pKa = 9.92DD114 pKa = 3.2YY115 pKa = 11.1GNPVHH120 pKa = 7.0FVPQDD125 pKa = 3.43VQRR128 pKa = 11.84KK129 pKa = 8.58GNADD133 pKa = 3.15FSKK136 pKa = 10.88NMKK139 pKa = 10.18AGTIGRR145 pKa = 11.84AYY147 pKa = 9.86IPLTAAATWAAYY159 pKa = 10.19YY160 pKa = 9.97PLGLKK165 pKa = 10.27ASYY168 pKa = 11.1NKK170 pKa = 8.87VQNYY174 pKa = 8.36GNPFLDD180 pKa = 4.01GANTILAWGGKK191 pKa = 8.9IDD193 pKa = 4.74GKK195 pKa = 10.89GGSPNSGSSDD205 pKa = 3.32SSSDD209 pKa = 3.31SGGNSLLALAKK220 pKa = 10.19QAMQEE225 pKa = 4.1LLKK228 pKa = 10.63KK229 pKa = 10.48VQDD232 pKa = 3.53ALQWDD237 pKa = 4.06VHH239 pKa = 7.39SIGSDD244 pKa = 2.68KK245 pKa = 10.79FFSNDD250 pKa = 2.89YY251 pKa = 8.37FTLQKK256 pKa = 9.76TFNNTYY262 pKa = 9.12HH263 pKa = 7.0IKK265 pKa = 8.04MTIGLLDD272 pKa = 3.83SLKK275 pKa = 10.93KK276 pKa = 10.89LIDD279 pKa = 3.73SVQVDD284 pKa = 3.66SGGSSSNPTDD294 pKa = 4.41DD295 pKa = 6.01DD296 pKa = 4.66GDD298 pKa = 3.93HH299 pKa = 6.69KK300 pKa = 10.7PISGKK305 pKa = 7.43SVKK308 pKa = 10.22PNGKK312 pKa = 8.68SGRR315 pKa = 11.84VIGGNWTYY323 pKa = 11.51AQLPEE328 pKa = 4.51KK329 pKa = 9.49YY330 pKa = 9.06KK331 pKa = 10.77KK332 pKa = 10.74AIGVPLFKK340 pKa = 10.35KK341 pKa = 10.27EE342 pKa = 3.9YY343 pKa = 9.69LYY345 pKa = 11.13KK346 pKa = 10.3PGNIFPQTGNAGQCTEE362 pKa = 4.3LTWAYY367 pKa = 9.46MSQLHH372 pKa = 6.84GKK374 pKa = 8.57RR375 pKa = 11.84QPTDD379 pKa = 3.37DD380 pKa = 3.88GQVTNGQRR388 pKa = 11.84VWYY391 pKa = 9.59VYY393 pKa = 10.68KK394 pKa = 10.88KK395 pKa = 10.51LGAKK399 pKa = 6.42TTHH402 pKa = 6.34NPTVGYY408 pKa = 9.76GFSSKK413 pKa = 10.35PPYY416 pKa = 10.08LQATAYY422 pKa = 10.12GIGHH426 pKa = 6.67TGVVVAVFDD435 pKa = 4.62DD436 pKa = 4.68GSFLVANYY444 pKa = 8.94NVPPYY449 pKa = 9.33VAPSRR454 pKa = 11.84VVLYY458 pKa = 8.72TLINGVPHH466 pKa = 6.57NAGDD470 pKa = 3.9NIVFFSGIAA479 pKa = 3.09
Molecular weight: 52.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5467 |
54 |
762 |
273.4 |
31.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.951 ± 0.42 | 0.512 ± 0.147 |
7.481 ± 0.354 | 6.292 ± 0.677 |
5.433 ± 0.389 | 4.701 ± 0.881 |
1.774 ± 0.222 | 7.353 ± 0.594 |
8.579 ± 0.364 | 7.92 ± 0.284 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.579 ± 0.292 | 8.652 ± 0.545 |
2.543 ± 0.325 | 3.933 ± 0.353 |
3.329 ± 0.294 | 6.311 ± 0.361 |
6.109 ± 0.426 | 5.396 ± 0.302 |
0.933 ± 0.222 | 6.219 ± 0.546 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
