
Millipede associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.39
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
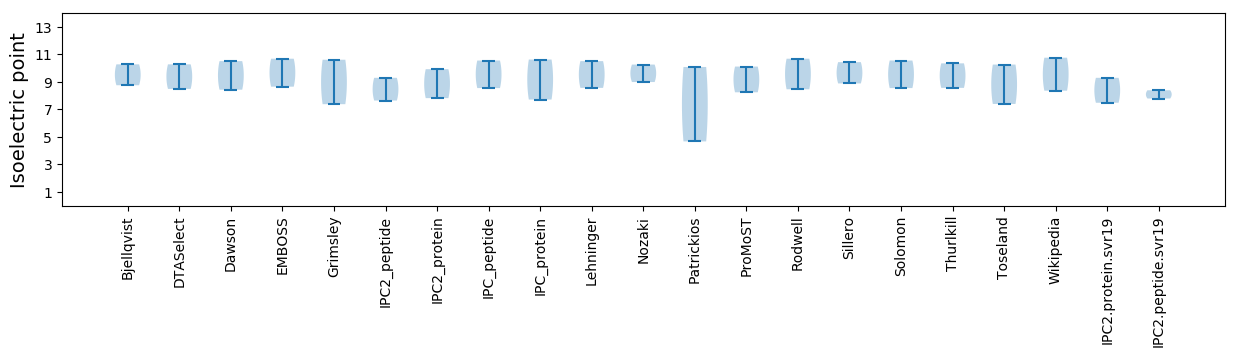
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A346BPC9|A0A346BPC9_9VIRU Putative capsid protein OS=Millipede associated circular virus 1 OX=2293296 PE=4 SV=1
MM1 pKa = 7.78PKK3 pKa = 10.65AKK5 pKa = 9.8FWCFTSFKK13 pKa = 10.44DD14 pKa = 3.41EE15 pKa = 4.42HH16 pKa = 7.03PNFCEE21 pKa = 3.99EE22 pKa = 4.04TFSYY26 pKa = 8.95ITYY29 pKa = 9.76QRR31 pKa = 11.84EE32 pKa = 3.89RR33 pKa = 11.84CPTSSRR39 pKa = 11.84EE40 pKa = 3.58HH41 pKa = 4.65WQGYY45 pKa = 6.87VEE47 pKa = 4.85CKK49 pKa = 9.72RR50 pKa = 11.84SSGVRR55 pKa = 11.84TVKK58 pKa = 10.83DD59 pKa = 3.51GLGDD63 pKa = 3.79NACHH67 pKa = 7.04LEE69 pKa = 4.09PSRR72 pKa = 11.84STATWFKK79 pKa = 8.79MARR82 pKa = 11.84IATLSKK88 pKa = 10.05SVRR91 pKa = 11.84RR92 pKa = 11.84GQRR95 pKa = 11.84TDD97 pKa = 3.12LEE99 pKa = 4.34RR100 pKa = 11.84VAKK103 pKa = 9.99RR104 pKa = 11.84VRR106 pKa = 11.84DD107 pKa = 3.79GGNLAEE113 pKa = 5.03IATEE117 pKa = 4.45FPVEE121 pKa = 4.12TIKK124 pKa = 10.75FGRR127 pKa = 11.84GIQQLIEE134 pKa = 4.25LNRR137 pKa = 11.84KK138 pKa = 8.56GRR140 pKa = 11.84DD141 pKa = 3.26GTIEE145 pKa = 4.13AKK147 pKa = 10.73VIIYY151 pKa = 8.58WGKK154 pKa = 7.92TGTGKK159 pKa = 8.02TRR161 pKa = 11.84RR162 pKa = 11.84AFEE165 pKa = 4.34RR166 pKa = 11.84FPTAYY171 pKa = 10.03FKK173 pKa = 10.8PDD175 pKa = 3.26GKK177 pKa = 10.12WFDD180 pKa = 3.66NYY182 pKa = 10.89NGEE185 pKa = 4.44DD186 pKa = 3.62VIIFDD191 pKa = 5.39DD192 pKa = 4.38FNGHH196 pKa = 6.21KK197 pKa = 10.29DD198 pKa = 3.31INVGMLLRR206 pKa = 11.84ICDD209 pKa = 4.43RR210 pKa = 11.84YY211 pKa = 10.72PMTVEE216 pKa = 4.93KK217 pKa = 10.6KK218 pKa = 10.78GSSCKK223 pKa = 10.52LNATTFVFTSNLHH236 pKa = 4.46WKK238 pKa = 8.57EE239 pKa = 3.76WYY241 pKa = 9.89NIEE244 pKa = 5.25GMRR247 pKa = 11.84QDD249 pKa = 5.31WIAHH253 pKa = 5.34WDD255 pKa = 3.3AFEE258 pKa = 4.14RR259 pKa = 11.84RR260 pKa = 11.84ITEE263 pKa = 3.56IEE265 pKa = 3.91AFDD268 pKa = 3.74AVEE271 pKa = 4.87PIDD274 pKa = 5.15NILIMM279 pKa = 4.75
MM1 pKa = 7.78PKK3 pKa = 10.65AKK5 pKa = 9.8FWCFTSFKK13 pKa = 10.44DD14 pKa = 3.41EE15 pKa = 4.42HH16 pKa = 7.03PNFCEE21 pKa = 3.99EE22 pKa = 4.04TFSYY26 pKa = 8.95ITYY29 pKa = 9.76QRR31 pKa = 11.84EE32 pKa = 3.89RR33 pKa = 11.84CPTSSRR39 pKa = 11.84EE40 pKa = 3.58HH41 pKa = 4.65WQGYY45 pKa = 6.87VEE47 pKa = 4.85CKK49 pKa = 9.72RR50 pKa = 11.84SSGVRR55 pKa = 11.84TVKK58 pKa = 10.83DD59 pKa = 3.51GLGDD63 pKa = 3.79NACHH67 pKa = 7.04LEE69 pKa = 4.09PSRR72 pKa = 11.84STATWFKK79 pKa = 8.79MARR82 pKa = 11.84IATLSKK88 pKa = 10.05SVRR91 pKa = 11.84RR92 pKa = 11.84GQRR95 pKa = 11.84TDD97 pKa = 3.12LEE99 pKa = 4.34RR100 pKa = 11.84VAKK103 pKa = 9.99RR104 pKa = 11.84VRR106 pKa = 11.84DD107 pKa = 3.79GGNLAEE113 pKa = 5.03IATEE117 pKa = 4.45FPVEE121 pKa = 4.12TIKK124 pKa = 10.75FGRR127 pKa = 11.84GIQQLIEE134 pKa = 4.25LNRR137 pKa = 11.84KK138 pKa = 8.56GRR140 pKa = 11.84DD141 pKa = 3.26GTIEE145 pKa = 4.13AKK147 pKa = 10.73VIIYY151 pKa = 8.58WGKK154 pKa = 7.92TGTGKK159 pKa = 8.02TRR161 pKa = 11.84RR162 pKa = 11.84AFEE165 pKa = 4.34RR166 pKa = 11.84FPTAYY171 pKa = 10.03FKK173 pKa = 10.8PDD175 pKa = 3.26GKK177 pKa = 10.12WFDD180 pKa = 3.66NYY182 pKa = 10.89NGEE185 pKa = 4.44DD186 pKa = 3.62VIIFDD191 pKa = 5.39DD192 pKa = 4.38FNGHH196 pKa = 6.21KK197 pKa = 10.29DD198 pKa = 3.31INVGMLLRR206 pKa = 11.84ICDD209 pKa = 4.43RR210 pKa = 11.84YY211 pKa = 10.72PMTVEE216 pKa = 4.93KK217 pKa = 10.6KK218 pKa = 10.78GSSCKK223 pKa = 10.52LNATTFVFTSNLHH236 pKa = 4.46WKK238 pKa = 8.57EE239 pKa = 3.76WYY241 pKa = 9.89NIEE244 pKa = 5.25GMRR247 pKa = 11.84QDD249 pKa = 5.31WIAHH253 pKa = 5.34WDD255 pKa = 3.3AFEE258 pKa = 4.14RR259 pKa = 11.84RR260 pKa = 11.84ITEE263 pKa = 3.56IEE265 pKa = 3.91AFDD268 pKa = 3.74AVEE271 pKa = 4.87PIDD274 pKa = 5.15NILIMM279 pKa = 4.75
Molecular weight: 32.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPC9|A0A346BPC9_9VIRU Putative capsid protein OS=Millipede associated circular virus 1 OX=2293296 PE=4 SV=1
MM1 pKa = 7.54RR2 pKa = 11.84RR3 pKa = 11.84YY4 pKa = 9.48SKK6 pKa = 10.53KK7 pKa = 9.53RR8 pKa = 11.84SYY10 pKa = 10.92RR11 pKa = 11.84SASRR15 pKa = 11.84SRR17 pKa = 11.84RR18 pKa = 11.84FRR20 pKa = 11.84KK21 pKa = 8.77GVRR24 pKa = 11.84RR25 pKa = 11.84TRR27 pKa = 11.84VRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84IRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SSKK39 pKa = 9.79GNYY42 pKa = 8.16FRR44 pKa = 11.84RR45 pKa = 11.84TFVDD49 pKa = 3.19VGSIIGTLGDD59 pKa = 4.76DD60 pKa = 4.01GTFSPLTPNSDD71 pKa = 3.24GTFTINTPGTFSIYY85 pKa = 10.91VKK87 pKa = 8.13TTILDD92 pKa = 4.31SIPTASSYY100 pKa = 11.27LLEE103 pKa = 4.27FEE105 pKa = 4.73AYY107 pKa = 9.65RR108 pKa = 11.84LRR110 pKa = 11.84WCKK113 pKa = 10.33LRR115 pKa = 11.84VRR117 pKa = 11.84NVTAQYY123 pKa = 11.17LVATNSGDD131 pKa = 4.34DD132 pKa = 3.63VTPKK136 pKa = 10.24QGSVYY141 pKa = 10.12YY142 pKa = 10.98VPDD145 pKa = 3.61KK146 pKa = 11.21YY147 pKa = 10.72RR148 pKa = 11.84IPSFSASSNYY158 pKa = 9.72IKK160 pKa = 10.63DD161 pKa = 3.1ILQYY165 pKa = 9.12MPKK168 pKa = 10.04FAHH171 pKa = 6.0QGQNFYY177 pKa = 11.22CFNRR181 pKa = 11.84NPKK184 pKa = 9.76YY185 pKa = 9.36LTPTISSDD193 pKa = 3.81SIGGNISQYY202 pKa = 11.15NSNLSCDD209 pKa = 3.92WIRR212 pKa = 11.84AQTTSGTQGTFPYY225 pKa = 9.98YY226 pKa = 10.1NLMWMVVDD234 pKa = 5.32SPASAQMQFRR244 pKa = 11.84FEE246 pKa = 5.73LEE248 pKa = 4.37TNWEE252 pKa = 4.38FKK254 pKa = 10.47GRR256 pKa = 11.84PHH258 pKa = 7.44GSYY261 pKa = 10.95AA262 pKa = 3.46
MM1 pKa = 7.54RR2 pKa = 11.84RR3 pKa = 11.84YY4 pKa = 9.48SKK6 pKa = 10.53KK7 pKa = 9.53RR8 pKa = 11.84SYY10 pKa = 10.92RR11 pKa = 11.84SASRR15 pKa = 11.84SRR17 pKa = 11.84RR18 pKa = 11.84FRR20 pKa = 11.84KK21 pKa = 8.77GVRR24 pKa = 11.84RR25 pKa = 11.84TRR27 pKa = 11.84VRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84IRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SSKK39 pKa = 9.79GNYY42 pKa = 8.16FRR44 pKa = 11.84RR45 pKa = 11.84TFVDD49 pKa = 3.19VGSIIGTLGDD59 pKa = 4.76DD60 pKa = 4.01GTFSPLTPNSDD71 pKa = 3.24GTFTINTPGTFSIYY85 pKa = 10.91VKK87 pKa = 8.13TTILDD92 pKa = 4.31SIPTASSYY100 pKa = 11.27LLEE103 pKa = 4.27FEE105 pKa = 4.73AYY107 pKa = 9.65RR108 pKa = 11.84LRR110 pKa = 11.84WCKK113 pKa = 10.33LRR115 pKa = 11.84VRR117 pKa = 11.84NVTAQYY123 pKa = 11.17LVATNSGDD131 pKa = 4.34DD132 pKa = 3.63VTPKK136 pKa = 10.24QGSVYY141 pKa = 10.12YY142 pKa = 10.98VPDD145 pKa = 3.61KK146 pKa = 11.21YY147 pKa = 10.72RR148 pKa = 11.84IPSFSASSNYY158 pKa = 9.72IKK160 pKa = 10.63DD161 pKa = 3.1ILQYY165 pKa = 9.12MPKK168 pKa = 10.04FAHH171 pKa = 6.0QGQNFYY177 pKa = 11.22CFNRR181 pKa = 11.84NPKK184 pKa = 9.76YY185 pKa = 9.36LTPTISSDD193 pKa = 3.81SIGGNISQYY202 pKa = 11.15NSNLSCDD209 pKa = 3.92WIRR212 pKa = 11.84AQTTSGTQGTFPYY225 pKa = 9.98YY226 pKa = 10.1NLMWMVVDD234 pKa = 5.32SPASAQMQFRR244 pKa = 11.84FEE246 pKa = 5.73LEE248 pKa = 4.37TNWEE252 pKa = 4.38FKK254 pKa = 10.47GRR256 pKa = 11.84PHH258 pKa = 7.44GSYY261 pKa = 10.95AA262 pKa = 3.46
Molecular weight: 30.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
541 |
262 |
279 |
270.5 |
31.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.991 ± 0.498 | 1.848 ± 0.442 |
5.36 ± 0.49 | 4.991 ± 1.937 |
6.1 ± 0.235 | 6.839 ± 0.22 |
1.479 ± 0.45 | 6.285 ± 0.592 |
5.915 ± 0.839 | 4.621 ± 0.214 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.078 | 4.806 ± 0.338 |
4.067 ± 0.563 | 2.957 ± 0.54 |
9.797 ± 0.799 | 7.948 ± 2.201 |
7.948 ± 0.282 | 4.806 ± 0.098 |
2.403 ± 0.551 | 4.806 ± 1.297 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
