
Sanxia sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
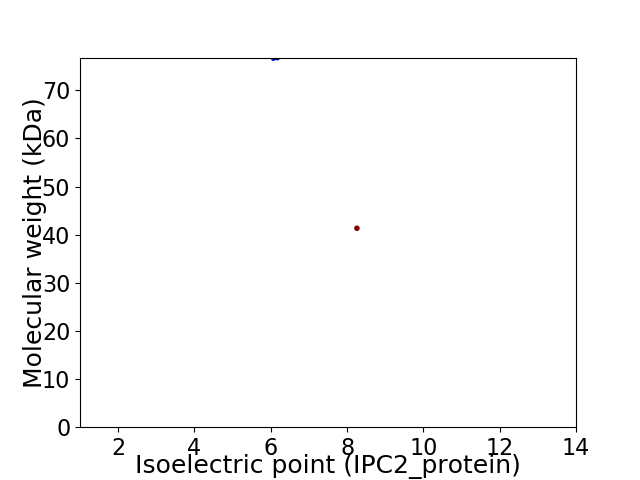
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
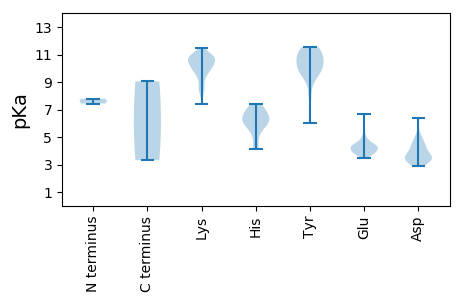
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEQ8|A0A1L3KEQ8_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 2 OX=1923381 PE=4 SV=1
MM1 pKa = 7.44SGSNYY6 pKa = 9.58TFLKK10 pKa = 10.63SALQLTTLLLCVGTFVDD27 pKa = 4.17TFDD30 pKa = 4.67SEE32 pKa = 4.54EE33 pKa = 4.4NKK35 pKa = 10.63CQMDD39 pKa = 4.34LNHH42 pKa = 7.07ALAAKK47 pKa = 9.69TEE49 pKa = 4.2MEE51 pKa = 4.46HH52 pKa = 4.78EE53 pKa = 4.21QKK55 pKa = 11.16YY56 pKa = 10.54LDD58 pKa = 3.76SKK60 pKa = 10.61MRR62 pKa = 11.84ALMDD66 pKa = 3.34KK67 pKa = 10.1MVEE70 pKa = 4.01EE71 pKa = 4.38KK72 pKa = 10.89AAVLLADD79 pKa = 3.79KK80 pKa = 10.8QILEE84 pKa = 4.31LAHH87 pKa = 6.49NLTVLGKK94 pKa = 7.38EE95 pKa = 4.09HH96 pKa = 7.16KK97 pKa = 10.18ALKK100 pKa = 10.22SKK102 pKa = 10.94QNLVEE107 pKa = 4.27RR108 pKa = 11.84DD109 pKa = 3.69FLKK112 pKa = 10.35MSRR115 pKa = 11.84KK116 pKa = 8.31LTGEE120 pKa = 3.76RR121 pKa = 11.84SRR123 pKa = 11.84NKK125 pKa = 10.24RR126 pKa = 11.84SITTKK131 pKa = 9.44EE132 pKa = 3.72QQIGNMIGKK141 pKa = 9.43SKK143 pKa = 9.62SQRR146 pKa = 11.84DD147 pKa = 3.55EE148 pKa = 4.86LEE150 pKa = 3.73EE151 pKa = 4.13CEE153 pKa = 4.21RR154 pKa = 11.84RR155 pKa = 11.84VIALQSSVDD164 pKa = 3.61SLNYY168 pKa = 9.61WKK170 pKa = 10.63DD171 pKa = 3.47EE172 pKa = 3.85LRR174 pKa = 11.84KK175 pKa = 9.94KK176 pKa = 10.69VEE178 pKa = 3.75QLEE181 pKa = 4.43DD182 pKa = 3.94DD183 pKa = 4.26NEE185 pKa = 4.11DD186 pKa = 3.05LRR188 pKa = 11.84EE189 pKa = 3.91RR190 pKa = 11.84MKK192 pKa = 10.88TSASQIDD199 pKa = 3.75FRR201 pKa = 11.84MVGGLILLFATVVVSLAWYY220 pKa = 8.45KK221 pKa = 10.01HH222 pKa = 5.44RR223 pKa = 11.84KK224 pKa = 8.68VVTLLKK230 pKa = 10.79EE231 pKa = 4.23EE232 pKa = 4.94IITLQTRR239 pKa = 11.84VEE241 pKa = 4.58SMTQRR246 pKa = 11.84YY247 pKa = 9.35SPVQMLANSMTNVSLGNSSNEE268 pKa = 3.96SLIPEE273 pKa = 4.41CKK275 pKa = 10.46VPGNALFPHH284 pKa = 7.14DD285 pKa = 5.04SDD287 pKa = 4.46PGQLTILLKK296 pKa = 10.7IGDD299 pKa = 4.37ALVAAGQVFWIKK311 pKa = 10.88DD312 pKa = 3.45WLVTAAHH319 pKa = 6.82VINAVSEE326 pKa = 4.3DD327 pKa = 3.52EE328 pKa = 5.62DD329 pKa = 5.0IILSSMVPQADD340 pKa = 3.79GTRR343 pKa = 11.84KK344 pKa = 8.47EE345 pKa = 3.92VRR347 pKa = 11.84VNRR350 pKa = 11.84SRR352 pKa = 11.84KK353 pKa = 9.81DD354 pKa = 3.35FLIPTPSFDD363 pKa = 3.7LAYY366 pKa = 9.88MKK368 pKa = 10.6VDD370 pKa = 3.32NSFMQQFKK378 pKa = 11.01SNKK381 pKa = 8.97FKK383 pKa = 10.78TNHH386 pKa = 6.34LSSDD390 pKa = 3.92VYY392 pKa = 11.26VSVSAEE398 pKa = 3.64GMASTGHH405 pKa = 6.02MEE407 pKa = 4.36PNDD410 pKa = 4.49DD411 pKa = 4.93GSVTYY416 pKa = 10.58YY417 pKa = 11.31GSTTPGFSGAPYY429 pKa = 9.84LWDD432 pKa = 3.63GKK434 pKa = 10.11VVGMHH439 pKa = 6.48LGARR443 pKa = 11.84AKK445 pKa = 9.57EE446 pKa = 4.04VCQYY450 pKa = 10.33GLSIAIIEE458 pKa = 4.65RR459 pKa = 11.84ILTRR463 pKa = 11.84FSRR466 pKa = 11.84PYY468 pKa = 9.79KK469 pKa = 10.65AEE471 pKa = 3.89STEE474 pKa = 4.53DD475 pKa = 3.3EE476 pKa = 4.35LFEE479 pKa = 4.96KK480 pKa = 10.68NFYY483 pKa = 7.7EE484 pKa = 4.24TLEE487 pKa = 3.92EE488 pKa = 3.81RR489 pKa = 11.84AMVGGRR495 pKa = 11.84IKK497 pKa = 10.74AKK499 pKa = 10.18RR500 pKa = 11.84INPEE504 pKa = 3.91DD505 pKa = 3.34FVVYY509 pKa = 10.85YY510 pKa = 10.31KK511 pKa = 10.67DD512 pKa = 2.92KK513 pKa = 11.11HH514 pKa = 6.53GNHH517 pKa = 5.58QSGVISDD524 pKa = 4.09MDD526 pKa = 3.72IEE528 pKa = 6.68DD529 pKa = 4.32DD530 pKa = 5.47DD531 pKa = 4.27IYY533 pKa = 11.53NYY535 pKa = 11.0LDD537 pKa = 3.38FGGDD541 pKa = 3.31LEE543 pKa = 4.73EE544 pKa = 5.87ADD546 pKa = 3.82EE547 pKa = 4.78YY548 pKa = 11.0YY549 pKa = 10.93SKK551 pKa = 11.11RR552 pKa = 11.84NPKK555 pKa = 10.59DD556 pKa = 3.08SDD558 pKa = 3.23FRR560 pKa = 11.84KK561 pKa = 10.1RR562 pKa = 11.84KK563 pKa = 8.27GRR565 pKa = 11.84RR566 pKa = 11.84YY567 pKa = 10.15DD568 pKa = 3.27PEE570 pKa = 4.3VYY572 pKa = 10.34FDD574 pKa = 3.99VPAHH578 pKa = 6.0FPKK581 pKa = 10.51NSKK584 pKa = 10.08KK585 pKa = 8.76EE586 pKa = 3.55RR587 pKa = 11.84SMISRR592 pKa = 11.84EE593 pKa = 3.84MGKK596 pKa = 10.35KK597 pKa = 8.87IVEE600 pKa = 4.25VQTEE604 pKa = 4.0KK605 pKa = 11.05DD606 pKa = 3.43VEE608 pKa = 4.39EE609 pKa = 4.41NGKK612 pKa = 9.39IDD614 pKa = 4.16PAGDD618 pKa = 3.5AQKK621 pKa = 11.09QSIQKK626 pKa = 10.44SNDD629 pKa = 2.93PMISILAAMTKK640 pKa = 9.97ISDD643 pKa = 3.89GQEE646 pKa = 3.65SMLALLKK653 pKa = 10.48RR654 pKa = 11.84ALPSTSSGSKK664 pKa = 9.75RR665 pKa = 11.84GRR667 pKa = 11.84QSRR670 pKa = 11.84SPMSQNKK677 pKa = 9.05
MM1 pKa = 7.44SGSNYY6 pKa = 9.58TFLKK10 pKa = 10.63SALQLTTLLLCVGTFVDD27 pKa = 4.17TFDD30 pKa = 4.67SEE32 pKa = 4.54EE33 pKa = 4.4NKK35 pKa = 10.63CQMDD39 pKa = 4.34LNHH42 pKa = 7.07ALAAKK47 pKa = 9.69TEE49 pKa = 4.2MEE51 pKa = 4.46HH52 pKa = 4.78EE53 pKa = 4.21QKK55 pKa = 11.16YY56 pKa = 10.54LDD58 pKa = 3.76SKK60 pKa = 10.61MRR62 pKa = 11.84ALMDD66 pKa = 3.34KK67 pKa = 10.1MVEE70 pKa = 4.01EE71 pKa = 4.38KK72 pKa = 10.89AAVLLADD79 pKa = 3.79KK80 pKa = 10.8QILEE84 pKa = 4.31LAHH87 pKa = 6.49NLTVLGKK94 pKa = 7.38EE95 pKa = 4.09HH96 pKa = 7.16KK97 pKa = 10.18ALKK100 pKa = 10.22SKK102 pKa = 10.94QNLVEE107 pKa = 4.27RR108 pKa = 11.84DD109 pKa = 3.69FLKK112 pKa = 10.35MSRR115 pKa = 11.84KK116 pKa = 8.31LTGEE120 pKa = 3.76RR121 pKa = 11.84SRR123 pKa = 11.84NKK125 pKa = 10.24RR126 pKa = 11.84SITTKK131 pKa = 9.44EE132 pKa = 3.72QQIGNMIGKK141 pKa = 9.43SKK143 pKa = 9.62SQRR146 pKa = 11.84DD147 pKa = 3.55EE148 pKa = 4.86LEE150 pKa = 3.73EE151 pKa = 4.13CEE153 pKa = 4.21RR154 pKa = 11.84RR155 pKa = 11.84VIALQSSVDD164 pKa = 3.61SLNYY168 pKa = 9.61WKK170 pKa = 10.63DD171 pKa = 3.47EE172 pKa = 3.85LRR174 pKa = 11.84KK175 pKa = 9.94KK176 pKa = 10.69VEE178 pKa = 3.75QLEE181 pKa = 4.43DD182 pKa = 3.94DD183 pKa = 4.26NEE185 pKa = 4.11DD186 pKa = 3.05LRR188 pKa = 11.84EE189 pKa = 3.91RR190 pKa = 11.84MKK192 pKa = 10.88TSASQIDD199 pKa = 3.75FRR201 pKa = 11.84MVGGLILLFATVVVSLAWYY220 pKa = 8.45KK221 pKa = 10.01HH222 pKa = 5.44RR223 pKa = 11.84KK224 pKa = 8.68VVTLLKK230 pKa = 10.79EE231 pKa = 4.23EE232 pKa = 4.94IITLQTRR239 pKa = 11.84VEE241 pKa = 4.58SMTQRR246 pKa = 11.84YY247 pKa = 9.35SPVQMLANSMTNVSLGNSSNEE268 pKa = 3.96SLIPEE273 pKa = 4.41CKK275 pKa = 10.46VPGNALFPHH284 pKa = 7.14DD285 pKa = 5.04SDD287 pKa = 4.46PGQLTILLKK296 pKa = 10.7IGDD299 pKa = 4.37ALVAAGQVFWIKK311 pKa = 10.88DD312 pKa = 3.45WLVTAAHH319 pKa = 6.82VINAVSEE326 pKa = 4.3DD327 pKa = 3.52EE328 pKa = 5.62DD329 pKa = 5.0IILSSMVPQADD340 pKa = 3.79GTRR343 pKa = 11.84KK344 pKa = 8.47EE345 pKa = 3.92VRR347 pKa = 11.84VNRR350 pKa = 11.84SRR352 pKa = 11.84KK353 pKa = 9.81DD354 pKa = 3.35FLIPTPSFDD363 pKa = 3.7LAYY366 pKa = 9.88MKK368 pKa = 10.6VDD370 pKa = 3.32NSFMQQFKK378 pKa = 11.01SNKK381 pKa = 8.97FKK383 pKa = 10.78TNHH386 pKa = 6.34LSSDD390 pKa = 3.92VYY392 pKa = 11.26VSVSAEE398 pKa = 3.64GMASTGHH405 pKa = 6.02MEE407 pKa = 4.36PNDD410 pKa = 4.49DD411 pKa = 4.93GSVTYY416 pKa = 10.58YY417 pKa = 11.31GSTTPGFSGAPYY429 pKa = 9.84LWDD432 pKa = 3.63GKK434 pKa = 10.11VVGMHH439 pKa = 6.48LGARR443 pKa = 11.84AKK445 pKa = 9.57EE446 pKa = 4.04VCQYY450 pKa = 10.33GLSIAIIEE458 pKa = 4.65RR459 pKa = 11.84ILTRR463 pKa = 11.84FSRR466 pKa = 11.84PYY468 pKa = 9.79KK469 pKa = 10.65AEE471 pKa = 3.89STEE474 pKa = 4.53DD475 pKa = 3.3EE476 pKa = 4.35LFEE479 pKa = 4.96KK480 pKa = 10.68NFYY483 pKa = 7.7EE484 pKa = 4.24TLEE487 pKa = 3.92EE488 pKa = 3.81RR489 pKa = 11.84AMVGGRR495 pKa = 11.84IKK497 pKa = 10.74AKK499 pKa = 10.18RR500 pKa = 11.84INPEE504 pKa = 3.91DD505 pKa = 3.34FVVYY509 pKa = 10.85YY510 pKa = 10.31KK511 pKa = 10.67DD512 pKa = 2.92KK513 pKa = 11.11HH514 pKa = 6.53GNHH517 pKa = 5.58QSGVISDD524 pKa = 4.09MDD526 pKa = 3.72IEE528 pKa = 6.68DD529 pKa = 4.32DD530 pKa = 5.47DD531 pKa = 4.27IYY533 pKa = 11.53NYY535 pKa = 11.0LDD537 pKa = 3.38FGGDD541 pKa = 3.31LEE543 pKa = 4.73EE544 pKa = 5.87ADD546 pKa = 3.82EE547 pKa = 4.78YY548 pKa = 11.0YY549 pKa = 10.93SKK551 pKa = 11.11RR552 pKa = 11.84NPKK555 pKa = 10.59DD556 pKa = 3.08SDD558 pKa = 3.23FRR560 pKa = 11.84KK561 pKa = 10.1RR562 pKa = 11.84KK563 pKa = 8.27GRR565 pKa = 11.84RR566 pKa = 11.84YY567 pKa = 10.15DD568 pKa = 3.27PEE570 pKa = 4.3VYY572 pKa = 10.34FDD574 pKa = 3.99VPAHH578 pKa = 6.0FPKK581 pKa = 10.51NSKK584 pKa = 10.08KK585 pKa = 8.76EE586 pKa = 3.55RR587 pKa = 11.84SMISRR592 pKa = 11.84EE593 pKa = 3.84MGKK596 pKa = 10.35KK597 pKa = 8.87IVEE600 pKa = 4.25VQTEE604 pKa = 4.0KK605 pKa = 11.05DD606 pKa = 3.43VEE608 pKa = 4.39EE609 pKa = 4.41NGKK612 pKa = 9.39IDD614 pKa = 4.16PAGDD618 pKa = 3.5AQKK621 pKa = 11.09QSIQKK626 pKa = 10.44SNDD629 pKa = 2.93PMISILAAMTKK640 pKa = 9.97ISDD643 pKa = 3.89GQEE646 pKa = 3.65SMLALLKK653 pKa = 10.48RR654 pKa = 11.84ALPSTSSGSKK664 pKa = 9.75RR665 pKa = 11.84GRR667 pKa = 11.84QSRR670 pKa = 11.84SPMSQNKK677 pKa = 9.05
Molecular weight: 76.69 kDa
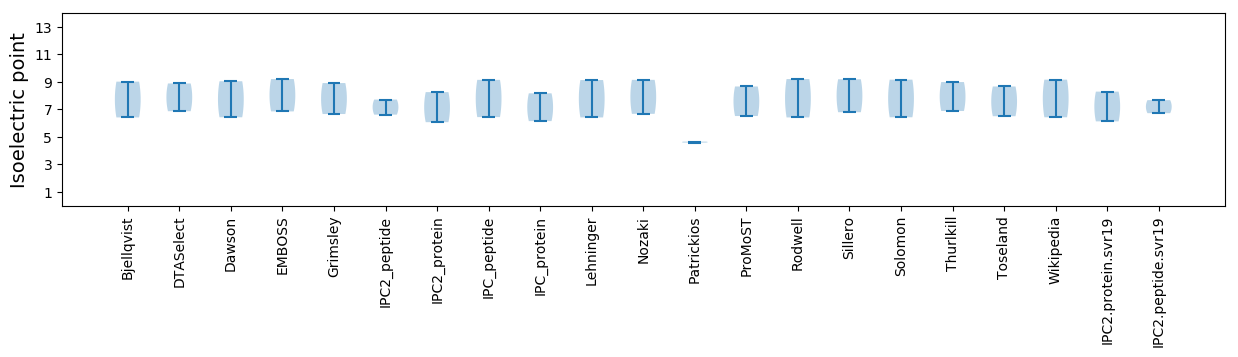
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEQ8|A0A1L3KEQ8_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 2 OX=1923381 PE=4 SV=1
MM1 pKa = 7.79DD2 pKa = 4.56VDD4 pKa = 4.86LSSIPGWCVLKK15 pKa = 11.01DD16 pKa = 3.37LGTTNRR22 pKa = 11.84EE23 pKa = 3.78VFKK26 pKa = 10.98VDD28 pKa = 2.99EE29 pKa = 4.42EE30 pKa = 4.36GRR32 pKa = 11.84YY33 pKa = 9.75NLDD36 pKa = 3.09RR37 pKa = 11.84VKK39 pKa = 10.34MVIEE43 pKa = 4.03IVKK46 pKa = 9.58KK47 pKa = 8.66RR48 pKa = 11.84WMEE51 pKa = 3.86IKK53 pKa = 10.46EE54 pKa = 4.16SDD56 pKa = 3.98PIYY59 pKa = 10.88AFVKK63 pKa = 9.98PEE65 pKa = 3.48PHH67 pKa = 6.79KK68 pKa = 10.74RR69 pKa = 11.84EE70 pKa = 3.96KK71 pKa = 10.97LNSGRR76 pKa = 11.84LRR78 pKa = 11.84LISGISLIDD87 pKa = 3.57SLIDD91 pKa = 3.06RR92 pKa = 11.84LLFMKK97 pKa = 8.21WTLKK101 pKa = 8.3TRR103 pKa = 11.84QMLTKK108 pKa = 10.25TPIAVGWTPLDD119 pKa = 3.26AVKK122 pKa = 9.89FHH124 pKa = 7.39HH125 pKa = 6.52MMGGMHH131 pKa = 6.29EE132 pKa = 4.42QYY134 pKa = 11.44LDD136 pKa = 3.44VDD138 pKa = 3.95KK139 pKa = 11.51SAWDD143 pKa = 3.38WSLKK147 pKa = 10.05PWILDD152 pKa = 3.11MVKK155 pKa = 10.16RR156 pKa = 11.84VLCGLVVAPTWWHH169 pKa = 4.13QRR171 pKa = 11.84VNKK174 pKa = 10.16RR175 pKa = 11.84FEE177 pKa = 4.34LLFGNPIWKK186 pKa = 9.22FQDD189 pKa = 2.88SSMIQQKK196 pKa = 9.79QPGIMKK202 pKa = 9.38SGCYY206 pKa = 6.02MTIWINSIGQLILHH220 pKa = 6.16QLAQSRR226 pKa = 11.84CGVQMHH232 pKa = 5.84VPMVVGDD239 pKa = 4.45DD240 pKa = 3.63TTQPYY245 pKa = 10.36YY246 pKa = 11.23GEE248 pKa = 5.46DD249 pKa = 2.95VDD251 pKa = 6.37AYY253 pKa = 10.88LQEE256 pKa = 3.94TRR258 pKa = 11.84NLGFVIKK265 pKa = 10.02PSIGRR270 pKa = 11.84IAEE273 pKa = 3.99FCGFRR278 pKa = 11.84FDD280 pKa = 4.72GFKK283 pKa = 10.8SLPAYY288 pKa = 9.93RR289 pKa = 11.84SKK291 pKa = 10.92HH292 pKa = 4.19QFLLRR297 pKa = 11.84HH298 pKa = 5.76LTLDD302 pKa = 4.92DD303 pKa = 5.12EE304 pKa = 5.03IATQTIQSYY313 pKa = 10.02QLLYY317 pKa = 11.35YY318 pKa = 9.37NDD320 pKa = 4.36KK321 pKa = 11.04YY322 pKa = 11.45ALQQIRR328 pKa = 11.84TLAIQRR334 pKa = 11.84GLLKK338 pKa = 10.63AVVPDD343 pKa = 3.56HH344 pKa = 6.11VLWLIIDD351 pKa = 4.61GLQTT355 pKa = 3.32
MM1 pKa = 7.79DD2 pKa = 4.56VDD4 pKa = 4.86LSSIPGWCVLKK15 pKa = 11.01DD16 pKa = 3.37LGTTNRR22 pKa = 11.84EE23 pKa = 3.78VFKK26 pKa = 10.98VDD28 pKa = 2.99EE29 pKa = 4.42EE30 pKa = 4.36GRR32 pKa = 11.84YY33 pKa = 9.75NLDD36 pKa = 3.09RR37 pKa = 11.84VKK39 pKa = 10.34MVIEE43 pKa = 4.03IVKK46 pKa = 9.58KK47 pKa = 8.66RR48 pKa = 11.84WMEE51 pKa = 3.86IKK53 pKa = 10.46EE54 pKa = 4.16SDD56 pKa = 3.98PIYY59 pKa = 10.88AFVKK63 pKa = 9.98PEE65 pKa = 3.48PHH67 pKa = 6.79KK68 pKa = 10.74RR69 pKa = 11.84EE70 pKa = 3.96KK71 pKa = 10.97LNSGRR76 pKa = 11.84LRR78 pKa = 11.84LISGISLIDD87 pKa = 3.57SLIDD91 pKa = 3.06RR92 pKa = 11.84LLFMKK97 pKa = 8.21WTLKK101 pKa = 8.3TRR103 pKa = 11.84QMLTKK108 pKa = 10.25TPIAVGWTPLDD119 pKa = 3.26AVKK122 pKa = 9.89FHH124 pKa = 7.39HH125 pKa = 6.52MMGGMHH131 pKa = 6.29EE132 pKa = 4.42QYY134 pKa = 11.44LDD136 pKa = 3.44VDD138 pKa = 3.95KK139 pKa = 11.51SAWDD143 pKa = 3.38WSLKK147 pKa = 10.05PWILDD152 pKa = 3.11MVKK155 pKa = 10.16RR156 pKa = 11.84VLCGLVVAPTWWHH169 pKa = 4.13QRR171 pKa = 11.84VNKK174 pKa = 10.16RR175 pKa = 11.84FEE177 pKa = 4.34LLFGNPIWKK186 pKa = 9.22FQDD189 pKa = 2.88SSMIQQKK196 pKa = 9.79QPGIMKK202 pKa = 9.38SGCYY206 pKa = 6.02MTIWINSIGQLILHH220 pKa = 6.16QLAQSRR226 pKa = 11.84CGVQMHH232 pKa = 5.84VPMVVGDD239 pKa = 4.45DD240 pKa = 3.63TTQPYY245 pKa = 10.36YY246 pKa = 11.23GEE248 pKa = 5.46DD249 pKa = 2.95VDD251 pKa = 6.37AYY253 pKa = 10.88LQEE256 pKa = 3.94TRR258 pKa = 11.84NLGFVIKK265 pKa = 10.02PSIGRR270 pKa = 11.84IAEE273 pKa = 3.99FCGFRR278 pKa = 11.84FDD280 pKa = 4.72GFKK283 pKa = 10.8SLPAYY288 pKa = 9.93RR289 pKa = 11.84SKK291 pKa = 10.92HH292 pKa = 4.19QFLLRR297 pKa = 11.84HH298 pKa = 5.76LTLDD302 pKa = 4.92DD303 pKa = 5.12EE304 pKa = 5.03IATQTIQSYY313 pKa = 10.02QLLYY317 pKa = 11.35YY318 pKa = 9.37NDD320 pKa = 4.36KK321 pKa = 11.04YY322 pKa = 11.45ALQQIRR328 pKa = 11.84TLAIQRR334 pKa = 11.84GLLKK338 pKa = 10.63AVVPDD343 pKa = 3.56HH344 pKa = 6.11VLWLIIDD351 pKa = 4.61GLQTT355 pKa = 3.32
Molecular weight: 41.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1032 |
355 |
677 |
516.0 |
59.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.233 ± 0.868 | 0.969 ± 0.243 |
6.977 ± 0.119 | 6.492 ± 1.408 |
3.391 ± 0.149 | 5.717 ± 0.265 |
2.229 ± 0.325 | 5.814 ± 0.99 |
8.14 ± 0.606 | 9.593 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.973 ± 0.016 | 3.585 ± 0.736 |
3.488 ± 0.407 | 4.748 ± 0.645 |
5.62 ± 0.008 | 7.752 ± 1.482 |
4.748 ± 0.022 | 6.686 ± 0.197 |
1.647 ± 0.957 | 3.198 ± 0.101 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
