
Jurona vesiculovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Vesiculovirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
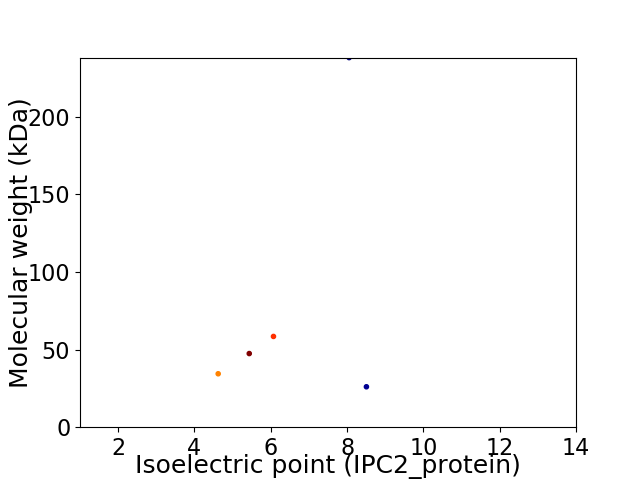
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1SV84|I1SV84_9RHAB Glycoprotein OS=Jurona vesiculovirus OX=1972568 PE=3 SV=1
MM1 pKa = 7.59KK2 pKa = 10.65KK3 pKa = 9.09MLTGINMSALKK14 pKa = 10.33NVLKK18 pKa = 10.53SYY20 pKa = 10.83PNLKK24 pKa = 8.46QTFDD28 pKa = 3.8EE29 pKa = 4.4LNEE32 pKa = 3.99MEE34 pKa = 4.66EE35 pKa = 4.29NGSVLQNPTNEE46 pKa = 4.33DD47 pKa = 3.18VSVEE51 pKa = 4.15HH52 pKa = 7.03PPLYY56 pKa = 10.38HH57 pKa = 6.96SLDD60 pKa = 3.44ILSEE64 pKa = 4.07SSEE67 pKa = 3.87EE68 pKa = 4.01DD69 pKa = 3.33TEE71 pKa = 4.35EE72 pKa = 3.99EE73 pKa = 5.18SEE75 pKa = 4.49VEE77 pKa = 4.77DD78 pKa = 3.94LGEE81 pKa = 4.06MEE83 pKa = 4.74NVSVDD88 pKa = 3.71DD89 pKa = 4.93EE90 pKa = 4.62KK91 pKa = 11.58DD92 pKa = 3.25QSEE95 pKa = 5.05EE96 pKa = 3.82IDD98 pKa = 3.71DD99 pKa = 5.17DD100 pKa = 4.02NFHH103 pKa = 7.21VSFNEE108 pKa = 3.83KK109 pKa = 10.61LPWTAMTQKK118 pKa = 9.19TVNGQLRR125 pKa = 11.84VNMSAPEE132 pKa = 4.05GLNEE136 pKa = 4.04LQFEE140 pKa = 4.38QWVSSIEE147 pKa = 3.94NLMSLSKK154 pKa = 10.48SLRR157 pKa = 11.84LHH159 pKa = 5.69SAEE162 pKa = 4.7LSIIEE167 pKa = 5.09DD168 pKa = 4.88GIQIDD173 pKa = 4.15EE174 pKa = 4.73KK175 pKa = 11.4LNSCLSRR182 pKa = 11.84TSCFKK187 pKa = 10.85ALPEE191 pKa = 4.36FKK193 pKa = 10.69DD194 pKa = 3.3PAGEE198 pKa = 4.2KK199 pKa = 10.08KK200 pKa = 10.65GSEE203 pKa = 4.23NLTIASSPSGSCEE216 pKa = 4.04SEE218 pKa = 4.08SSDD221 pKa = 4.76LSGLPSLHH229 pKa = 6.24QEE231 pKa = 4.2EE232 pKa = 4.62IKK234 pKa = 11.35NMIEE238 pKa = 3.73EE239 pKa = 4.2KK240 pKa = 10.54FILRR244 pKa = 11.84SEE246 pKa = 4.13DD247 pKa = 3.63KK248 pKa = 10.58KK249 pKa = 10.81CKK251 pKa = 9.77EE252 pKa = 3.89YY253 pKa = 8.89HH254 pKa = 5.6TSFAEE259 pKa = 4.06LFGSKK264 pKa = 9.04EE265 pKa = 3.53AALAFLNGNPVGIDD279 pKa = 3.39EE280 pKa = 4.78LVCAGLKK287 pKa = 10.05RR288 pKa = 11.84RR289 pKa = 11.84GIFNRR294 pKa = 11.84MRR296 pKa = 11.84IKK298 pKa = 10.83YY299 pKa = 8.23VLKK302 pKa = 10.48PIFKK306 pKa = 10.48
MM1 pKa = 7.59KK2 pKa = 10.65KK3 pKa = 9.09MLTGINMSALKK14 pKa = 10.33NVLKK18 pKa = 10.53SYY20 pKa = 10.83PNLKK24 pKa = 8.46QTFDD28 pKa = 3.8EE29 pKa = 4.4LNEE32 pKa = 3.99MEE34 pKa = 4.66EE35 pKa = 4.29NGSVLQNPTNEE46 pKa = 4.33DD47 pKa = 3.18VSVEE51 pKa = 4.15HH52 pKa = 7.03PPLYY56 pKa = 10.38HH57 pKa = 6.96SLDD60 pKa = 3.44ILSEE64 pKa = 4.07SSEE67 pKa = 3.87EE68 pKa = 4.01DD69 pKa = 3.33TEE71 pKa = 4.35EE72 pKa = 3.99EE73 pKa = 5.18SEE75 pKa = 4.49VEE77 pKa = 4.77DD78 pKa = 3.94LGEE81 pKa = 4.06MEE83 pKa = 4.74NVSVDD88 pKa = 3.71DD89 pKa = 4.93EE90 pKa = 4.62KK91 pKa = 11.58DD92 pKa = 3.25QSEE95 pKa = 5.05EE96 pKa = 3.82IDD98 pKa = 3.71DD99 pKa = 5.17DD100 pKa = 4.02NFHH103 pKa = 7.21VSFNEE108 pKa = 3.83KK109 pKa = 10.61LPWTAMTQKK118 pKa = 9.19TVNGQLRR125 pKa = 11.84VNMSAPEE132 pKa = 4.05GLNEE136 pKa = 4.04LQFEE140 pKa = 4.38QWVSSIEE147 pKa = 3.94NLMSLSKK154 pKa = 10.48SLRR157 pKa = 11.84LHH159 pKa = 5.69SAEE162 pKa = 4.7LSIIEE167 pKa = 5.09DD168 pKa = 4.88GIQIDD173 pKa = 4.15EE174 pKa = 4.73KK175 pKa = 11.4LNSCLSRR182 pKa = 11.84TSCFKK187 pKa = 10.85ALPEE191 pKa = 4.36FKK193 pKa = 10.69DD194 pKa = 3.3PAGEE198 pKa = 4.2KK199 pKa = 10.08KK200 pKa = 10.65GSEE203 pKa = 4.23NLTIASSPSGSCEE216 pKa = 4.04SEE218 pKa = 4.08SSDD221 pKa = 4.76LSGLPSLHH229 pKa = 6.24QEE231 pKa = 4.2EE232 pKa = 4.62IKK234 pKa = 11.35NMIEE238 pKa = 3.73EE239 pKa = 4.2KK240 pKa = 10.54FILRR244 pKa = 11.84SEE246 pKa = 4.13DD247 pKa = 3.63KK248 pKa = 10.58KK249 pKa = 10.81CKK251 pKa = 9.77EE252 pKa = 3.89YY253 pKa = 8.89HH254 pKa = 5.6TSFAEE259 pKa = 4.06LFGSKK264 pKa = 9.04EE265 pKa = 3.53AALAFLNGNPVGIDD279 pKa = 3.39EE280 pKa = 4.78LVCAGLKK287 pKa = 10.05RR288 pKa = 11.84RR289 pKa = 11.84GIFNRR294 pKa = 11.84MRR296 pKa = 11.84IKK298 pKa = 10.83YY299 pKa = 8.23VLKK302 pKa = 10.48PIFKK306 pKa = 10.48
Molecular weight: 34.42 kDa
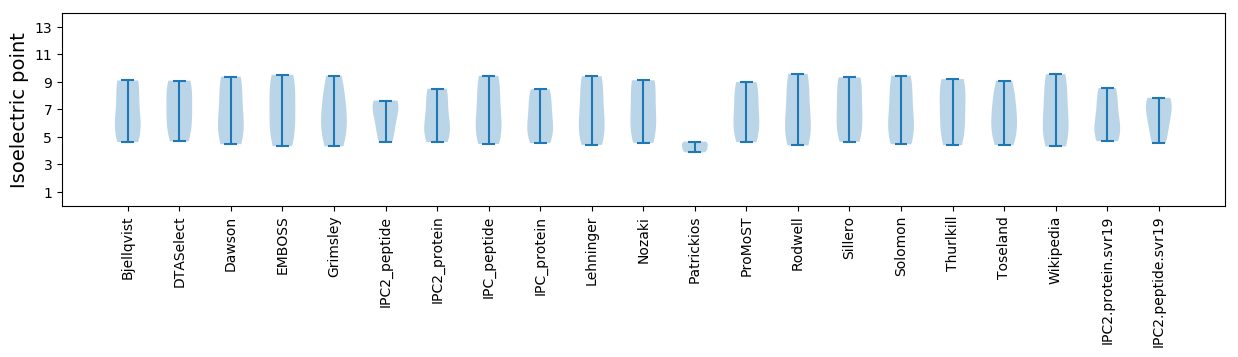
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1SV83|I1SV83_9RHAB Phosphoprotein OS=Jurona vesiculovirus OX=1972568 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.25SFKK5 pKa = 11.01GMFKK9 pKa = 10.56NYY11 pKa = 9.8KK12 pKa = 9.38EE13 pKa = 3.99KK14 pKa = 11.36NKK16 pKa = 9.99VKK18 pKa = 10.81KK19 pKa = 10.5EE20 pKa = 4.01MDD22 pKa = 3.04WDD24 pKa = 4.13SPPSYY29 pKa = 9.88TDD31 pKa = 2.95VRR33 pKa = 11.84RR34 pKa = 11.84GIYY37 pKa = 9.85PSAPLFGLDD46 pKa = 3.61DD47 pKa = 4.3SFMEE51 pKa = 4.45TLPSLGIQNMKK62 pKa = 10.16LQYY65 pKa = 9.88KK66 pKa = 10.35CSIQLRR72 pKa = 11.84AEE74 pKa = 4.34FPFVSYY80 pKa = 11.1SEE82 pKa = 4.4VVMALSQWDD91 pKa = 3.5EE92 pKa = 4.15EE93 pKa = 4.5YY94 pKa = 10.87RR95 pKa = 11.84GFLGKK100 pKa = 10.27RR101 pKa = 11.84PFYY104 pKa = 10.37RR105 pKa = 11.84AIILRR110 pKa = 11.84TSKK113 pKa = 9.25TLKK116 pKa = 10.36AVPLSLTDD124 pKa = 3.49GGRR127 pKa = 11.84PEE129 pKa = 4.34YY130 pKa = 10.6NGEE133 pKa = 3.92IQGQSSLYY141 pKa = 9.34HH142 pKa = 5.95SLGIIPPMMYY152 pKa = 10.19VPEE155 pKa = 4.22TFMKK159 pKa = 9.72EE160 pKa = 3.21WKK162 pKa = 8.28TAGNRR167 pKa = 11.84GILIIKK173 pKa = 8.98IWLGITDD180 pKa = 4.33TLDD183 pKa = 3.33TLDD186 pKa = 4.68PLLHH190 pKa = 6.11PMIFKK195 pKa = 10.26SEE197 pKa = 4.11RR198 pKa = 11.84EE199 pKa = 4.18LVEE202 pKa = 3.63MAKK205 pKa = 10.21VFNLDD210 pKa = 2.92ISKK213 pKa = 10.55GRR215 pKa = 11.84DD216 pKa = 3.33NNWIISRR223 pKa = 11.84SYY225 pKa = 10.86
MM1 pKa = 7.62KK2 pKa = 10.25SFKK5 pKa = 11.01GMFKK9 pKa = 10.56NYY11 pKa = 9.8KK12 pKa = 9.38EE13 pKa = 3.99KK14 pKa = 11.36NKK16 pKa = 9.99VKK18 pKa = 10.81KK19 pKa = 10.5EE20 pKa = 4.01MDD22 pKa = 3.04WDD24 pKa = 4.13SPPSYY29 pKa = 9.88TDD31 pKa = 2.95VRR33 pKa = 11.84RR34 pKa = 11.84GIYY37 pKa = 9.85PSAPLFGLDD46 pKa = 3.61DD47 pKa = 4.3SFMEE51 pKa = 4.45TLPSLGIQNMKK62 pKa = 10.16LQYY65 pKa = 9.88KK66 pKa = 10.35CSIQLRR72 pKa = 11.84AEE74 pKa = 4.34FPFVSYY80 pKa = 11.1SEE82 pKa = 4.4VVMALSQWDD91 pKa = 3.5EE92 pKa = 4.15EE93 pKa = 4.5YY94 pKa = 10.87RR95 pKa = 11.84GFLGKK100 pKa = 10.27RR101 pKa = 11.84PFYY104 pKa = 10.37RR105 pKa = 11.84AIILRR110 pKa = 11.84TSKK113 pKa = 9.25TLKK116 pKa = 10.36AVPLSLTDD124 pKa = 3.49GGRR127 pKa = 11.84PEE129 pKa = 4.34YY130 pKa = 10.6NGEE133 pKa = 3.92IQGQSSLYY141 pKa = 9.34HH142 pKa = 5.95SLGIIPPMMYY152 pKa = 10.19VPEE155 pKa = 4.22TFMKK159 pKa = 9.72EE160 pKa = 3.21WKK162 pKa = 8.28TAGNRR167 pKa = 11.84GILIIKK173 pKa = 8.98IWLGITDD180 pKa = 4.33TLDD183 pKa = 3.33TLDD186 pKa = 4.68PLLHH190 pKa = 6.11PMIFKK195 pKa = 10.26SEE197 pKa = 4.11RR198 pKa = 11.84EE199 pKa = 4.18LVEE202 pKa = 3.63MAKK205 pKa = 10.21VFNLDD210 pKa = 2.92ISKK213 pKa = 10.55GRR215 pKa = 11.84DD216 pKa = 3.33NNWIISRR223 pKa = 11.84SYY225 pKa = 10.86
Molecular weight: 26.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3567 |
225 |
2090 |
713.4 |
80.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.486 ± 0.67 | 1.71 ± 0.398 |
5.719 ± 0.322 | 6.224 ± 1.135 |
4.093 ± 0.239 | 6.28 ± 0.431 |
2.467 ± 0.363 | 6.869 ± 0.495 |
6.897 ± 0.33 | 10.177 ± 0.769 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.747 ± 0.313 | 4.878 ± 0.392 |
4.429 ± 0.556 | 3.196 ± 0.152 |
4.99 ± 0.637 | 9.532 ± 0.697 |
5.018 ± 0.438 | 5.13 ± 0.462 |
1.99 ± 0.239 | 3.168 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
