
Wenling frogfish arenavirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Arenaviridae; Antennavirus; Hairy antennavirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
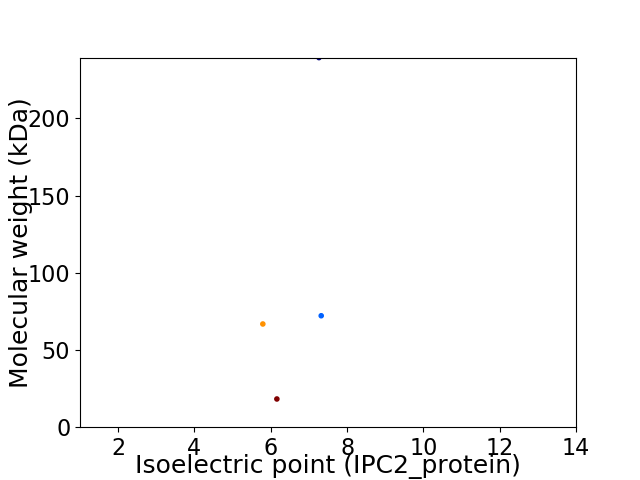
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
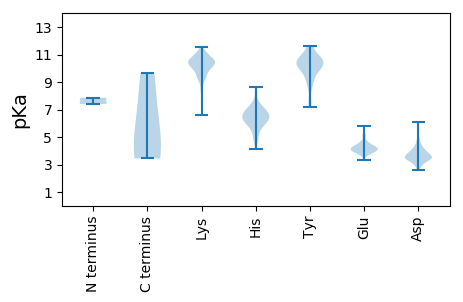
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2P1GNQ8|A0A2P1GNQ8_9VIRU Replicase OS=Wenling frogfish arenavirus 2 OX=2116467 PE=4 SV=1
MM1 pKa = 7.79AGTSDD6 pKa = 3.77EE7 pKa = 4.21NQLTFEE13 pKa = 4.55EE14 pKa = 4.8LKK16 pKa = 10.47QLHH19 pKa = 6.16GANHH23 pKa = 6.62VIDD26 pKa = 5.54DD27 pKa = 3.94NALVEE32 pKa = 4.24VKK34 pKa = 10.36IIFDD38 pKa = 4.08KK39 pKa = 11.31LDD41 pKa = 3.29GAIIKK46 pKa = 10.14EE47 pKa = 4.14LDD49 pKa = 3.66DD50 pKa = 3.44MSKK53 pKa = 10.71RR54 pKa = 11.84IAKK57 pKa = 10.03SEE59 pKa = 4.03LKK61 pKa = 10.59DD62 pKa = 4.0YY63 pKa = 10.99IDD65 pKa = 5.41DD66 pKa = 4.33LSDD69 pKa = 3.3VNRR72 pKa = 11.84KK73 pKa = 8.63IRR75 pKa = 11.84SLHH78 pKa = 4.59TSEE81 pKa = 4.48YY82 pKa = 9.91VSEE85 pKa = 5.05GSQEE89 pKa = 3.96PVQLSINEE97 pKa = 3.99LTAGEE102 pKa = 4.56VVQLDD107 pKa = 3.57QDD109 pKa = 3.7IEE111 pKa = 4.29DD112 pKa = 3.87MRR114 pKa = 11.84KK115 pKa = 9.82KK116 pKa = 10.83SNTGRR121 pKa = 11.84GTGNKK126 pKa = 9.68DD127 pKa = 2.63NWISGCEE134 pKa = 3.87KK135 pKa = 10.67DD136 pKa = 5.76LEE138 pKa = 4.44LLKK141 pKa = 10.46STGYY145 pKa = 11.17FDD147 pKa = 3.49MSKK150 pKa = 11.08GNPQSLANYY159 pKa = 8.59CLEE162 pKa = 4.32AGRR165 pKa = 11.84EE166 pKa = 4.12LEE168 pKa = 4.47LNQALEE174 pKa = 4.48DD175 pKa = 3.95SLILTDD181 pKa = 3.39EE182 pKa = 4.45DD183 pKa = 3.33KK184 pKa = 11.58ARR186 pKa = 11.84FVTEE190 pKa = 3.68KK191 pKa = 10.97RR192 pKa = 11.84EE193 pKa = 3.86GLAAANHH200 pKa = 5.75FQVDD204 pKa = 4.09PLFLLALFMKK214 pKa = 10.46ASQLTWQDD222 pKa = 3.25CVKK225 pKa = 10.63CITILKK231 pKa = 7.51TLLMEE236 pKa = 5.06RR237 pKa = 11.84PLSLKK242 pKa = 10.84LKK244 pKa = 9.95DD245 pKa = 3.81QLGKK249 pKa = 10.69KK250 pKa = 9.27PGGVFSFLNTLYY262 pKa = 10.39EE263 pKa = 4.44GKK265 pKa = 10.38LGLFGSPFIMTRR277 pKa = 11.84ARR279 pKa = 11.84LTYY282 pKa = 9.42CLSVMYY288 pKa = 8.76GIPSRR293 pKa = 11.84LAISASRR300 pKa = 11.84GVPSLIKK307 pKa = 10.77DD308 pKa = 3.36MLKK311 pKa = 9.91IRR313 pKa = 11.84ITKK316 pKa = 10.06KK317 pKa = 10.76LIISPDD323 pKa = 3.63RR324 pKa = 11.84EE325 pKa = 3.8DD326 pKa = 3.66STWYY330 pKa = 9.79EE331 pKa = 4.27HH332 pKa = 6.07GCHH335 pKa = 6.04RR336 pKa = 11.84VFTTPDD342 pKa = 3.55PEE344 pKa = 4.85DD345 pKa = 3.13VWKK348 pKa = 10.94NRR350 pKa = 11.84TAVVKK355 pKa = 10.69NGLSTVVINTKK366 pKa = 10.58ADD368 pKa = 3.36LRR370 pKa = 11.84SVKK373 pKa = 9.74EE374 pKa = 3.57VAAAPKK380 pKa = 10.06SSVRR384 pKa = 11.84RR385 pKa = 11.84SPEE388 pKa = 3.59NKK390 pKa = 7.88QTIAKK395 pKa = 8.12TSSHH399 pKa = 6.3LPIDD403 pKa = 3.63TFLVRR408 pKa = 11.84KK409 pKa = 7.66TPEE412 pKa = 4.14SQSSGPFWVIDD423 pKa = 3.59VEE425 pKa = 5.05GEE427 pKa = 3.9AKK429 pKa = 10.56SPVEE433 pKa = 3.57ICIMKK438 pKa = 9.92FDD440 pKa = 4.5PEE442 pKa = 4.11IGGKK446 pKa = 10.42GGIKK450 pKa = 10.26DD451 pKa = 3.34RR452 pKa = 11.84FFRR455 pKa = 11.84QIKK458 pKa = 10.35SNIGDD463 pKa = 3.38SSFTHH468 pKa = 6.79GLNKK472 pKa = 10.26SKK474 pKa = 10.87SRR476 pKa = 11.84DD477 pKa = 3.68DD478 pKa = 4.09VDD480 pKa = 3.22WADD483 pKa = 3.99DD484 pKa = 3.59MRR486 pKa = 11.84EE487 pKa = 3.79FWVKK491 pKa = 8.55VHH493 pKa = 6.27GNVYY497 pKa = 10.54CKK499 pKa = 10.44GSRR502 pKa = 11.84DD503 pKa = 3.47IKK505 pKa = 11.22DD506 pKa = 3.56FLDD509 pKa = 3.6NLGLSGTILDD519 pKa = 4.53LEE521 pKa = 5.2GIHH524 pKa = 7.05KK525 pKa = 10.54SWDD528 pKa = 4.21DD529 pKa = 3.53INKK532 pKa = 8.29TQYY535 pKa = 11.65ACILDD540 pKa = 4.06LEE542 pKa = 4.65STQVCSRR549 pKa = 11.84KK550 pKa = 10.07DD551 pKa = 3.15IHH553 pKa = 7.57DD554 pKa = 4.62KK555 pKa = 11.12LPQKK559 pKa = 10.91QNGDD563 pKa = 3.41INKK566 pKa = 9.31KK567 pKa = 9.68HH568 pKa = 6.36LPHH571 pKa = 6.61CAEE574 pKa = 4.13VDD576 pKa = 3.99CLHH579 pKa = 7.21LICMAMGKK587 pKa = 9.8VPDD590 pKa = 4.24EE591 pKa = 4.67FIKK594 pKa = 10.18TT595 pKa = 3.6
MM1 pKa = 7.79AGTSDD6 pKa = 3.77EE7 pKa = 4.21NQLTFEE13 pKa = 4.55EE14 pKa = 4.8LKK16 pKa = 10.47QLHH19 pKa = 6.16GANHH23 pKa = 6.62VIDD26 pKa = 5.54DD27 pKa = 3.94NALVEE32 pKa = 4.24VKK34 pKa = 10.36IIFDD38 pKa = 4.08KK39 pKa = 11.31LDD41 pKa = 3.29GAIIKK46 pKa = 10.14EE47 pKa = 4.14LDD49 pKa = 3.66DD50 pKa = 3.44MSKK53 pKa = 10.71RR54 pKa = 11.84IAKK57 pKa = 10.03SEE59 pKa = 4.03LKK61 pKa = 10.59DD62 pKa = 4.0YY63 pKa = 10.99IDD65 pKa = 5.41DD66 pKa = 4.33LSDD69 pKa = 3.3VNRR72 pKa = 11.84KK73 pKa = 8.63IRR75 pKa = 11.84SLHH78 pKa = 4.59TSEE81 pKa = 4.48YY82 pKa = 9.91VSEE85 pKa = 5.05GSQEE89 pKa = 3.96PVQLSINEE97 pKa = 3.99LTAGEE102 pKa = 4.56VVQLDD107 pKa = 3.57QDD109 pKa = 3.7IEE111 pKa = 4.29DD112 pKa = 3.87MRR114 pKa = 11.84KK115 pKa = 9.82KK116 pKa = 10.83SNTGRR121 pKa = 11.84GTGNKK126 pKa = 9.68DD127 pKa = 2.63NWISGCEE134 pKa = 3.87KK135 pKa = 10.67DD136 pKa = 5.76LEE138 pKa = 4.44LLKK141 pKa = 10.46STGYY145 pKa = 11.17FDD147 pKa = 3.49MSKK150 pKa = 11.08GNPQSLANYY159 pKa = 8.59CLEE162 pKa = 4.32AGRR165 pKa = 11.84EE166 pKa = 4.12LEE168 pKa = 4.47LNQALEE174 pKa = 4.48DD175 pKa = 3.95SLILTDD181 pKa = 3.39EE182 pKa = 4.45DD183 pKa = 3.33KK184 pKa = 11.58ARR186 pKa = 11.84FVTEE190 pKa = 3.68KK191 pKa = 10.97RR192 pKa = 11.84EE193 pKa = 3.86GLAAANHH200 pKa = 5.75FQVDD204 pKa = 4.09PLFLLALFMKK214 pKa = 10.46ASQLTWQDD222 pKa = 3.25CVKK225 pKa = 10.63CITILKK231 pKa = 7.51TLLMEE236 pKa = 5.06RR237 pKa = 11.84PLSLKK242 pKa = 10.84LKK244 pKa = 9.95DD245 pKa = 3.81QLGKK249 pKa = 10.69KK250 pKa = 9.27PGGVFSFLNTLYY262 pKa = 10.39EE263 pKa = 4.44GKK265 pKa = 10.38LGLFGSPFIMTRR277 pKa = 11.84ARR279 pKa = 11.84LTYY282 pKa = 9.42CLSVMYY288 pKa = 8.76GIPSRR293 pKa = 11.84LAISASRR300 pKa = 11.84GVPSLIKK307 pKa = 10.77DD308 pKa = 3.36MLKK311 pKa = 9.91IRR313 pKa = 11.84ITKK316 pKa = 10.06KK317 pKa = 10.76LIISPDD323 pKa = 3.63RR324 pKa = 11.84EE325 pKa = 3.8DD326 pKa = 3.66STWYY330 pKa = 9.79EE331 pKa = 4.27HH332 pKa = 6.07GCHH335 pKa = 6.04RR336 pKa = 11.84VFTTPDD342 pKa = 3.55PEE344 pKa = 4.85DD345 pKa = 3.13VWKK348 pKa = 10.94NRR350 pKa = 11.84TAVVKK355 pKa = 10.69NGLSTVVINTKK366 pKa = 10.58ADD368 pKa = 3.36LRR370 pKa = 11.84SVKK373 pKa = 9.74EE374 pKa = 3.57VAAAPKK380 pKa = 10.06SSVRR384 pKa = 11.84RR385 pKa = 11.84SPEE388 pKa = 3.59NKK390 pKa = 7.88QTIAKK395 pKa = 8.12TSSHH399 pKa = 6.3LPIDD403 pKa = 3.63TFLVRR408 pKa = 11.84KK409 pKa = 7.66TPEE412 pKa = 4.14SQSSGPFWVIDD423 pKa = 3.59VEE425 pKa = 5.05GEE427 pKa = 3.9AKK429 pKa = 10.56SPVEE433 pKa = 3.57ICIMKK438 pKa = 9.92FDD440 pKa = 4.5PEE442 pKa = 4.11IGGKK446 pKa = 10.42GGIKK450 pKa = 10.26DD451 pKa = 3.34RR452 pKa = 11.84FFRR455 pKa = 11.84QIKK458 pKa = 10.35SNIGDD463 pKa = 3.38SSFTHH468 pKa = 6.79GLNKK472 pKa = 10.26SKK474 pKa = 10.87SRR476 pKa = 11.84DD477 pKa = 3.68DD478 pKa = 4.09VDD480 pKa = 3.22WADD483 pKa = 3.99DD484 pKa = 3.59MRR486 pKa = 11.84EE487 pKa = 3.79FWVKK491 pKa = 8.55VHH493 pKa = 6.27GNVYY497 pKa = 10.54CKK499 pKa = 10.44GSRR502 pKa = 11.84DD503 pKa = 3.47IKK505 pKa = 11.22DD506 pKa = 3.56FLDD509 pKa = 3.6NLGLSGTILDD519 pKa = 4.53LEE521 pKa = 5.2GIHH524 pKa = 7.05KK525 pKa = 10.54SWDD528 pKa = 4.21DD529 pKa = 3.53INKK532 pKa = 8.29TQYY535 pKa = 11.65ACILDD540 pKa = 4.06LEE542 pKa = 4.65STQVCSRR549 pKa = 11.84KK550 pKa = 10.07DD551 pKa = 3.15IHH553 pKa = 7.57DD554 pKa = 4.62KK555 pKa = 11.12LPQKK559 pKa = 10.91QNGDD563 pKa = 3.41INKK566 pKa = 9.31KK567 pKa = 9.68HH568 pKa = 6.36LPHH571 pKa = 6.61CAEE574 pKa = 4.13VDD576 pKa = 3.99CLHH579 pKa = 7.21LICMAMGKK587 pKa = 9.8VPDD590 pKa = 4.24EE591 pKa = 4.67FIKK594 pKa = 10.18TT595 pKa = 3.6
Molecular weight: 66.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GNX5|A0A2P1GNX5_9VIRU Glycoprotein OS=Wenling frogfish arenavirus 2 OX=2116467 PE=4 SV=1
MM1 pKa = 7.44FEE3 pKa = 5.2SIIDD7 pKa = 4.85PILEE11 pKa = 3.88WLFNLFSFILAVIFVMTLSLLIYY34 pKa = 10.44RR35 pKa = 11.84MRR37 pKa = 11.84NLPFLFMMCGSAEE50 pKa = 4.03CWVSNIQMNGHH61 pKa = 4.84TAEE64 pKa = 3.87MRR66 pKa = 11.84QYY68 pKa = 10.44EE69 pKa = 4.39GQWLCGKK76 pKa = 10.14IKK78 pKa = 9.88MIKK81 pKa = 10.22LDD83 pKa = 3.57DD84 pKa = 4.23LYY86 pKa = 11.16YY87 pKa = 10.15IRR89 pKa = 11.84YY90 pKa = 9.18NMSDD94 pKa = 3.55SAVHH98 pKa = 6.47LVMSRR103 pKa = 11.84CTNTKK108 pKa = 10.34LEE110 pKa = 4.43NGWEE114 pKa = 4.66SVCSEE119 pKa = 4.34RR120 pKa = 11.84PALLKK125 pKa = 10.85NISTGISMIAGFNCSGQQKK144 pKa = 10.06CFVGDD149 pKa = 3.89GCTPWVDD156 pKa = 3.45ARR158 pKa = 11.84DD159 pKa = 3.3ICGRR163 pKa = 11.84KK164 pKa = 9.56LEE166 pKa = 4.47RR167 pKa = 11.84KK168 pKa = 9.49LDD170 pKa = 3.59NHH172 pKa = 6.56QMLDD176 pKa = 2.92ICRR179 pKa = 11.84QPIVGWIDD187 pKa = 3.17SDD189 pKa = 4.2GVHH192 pKa = 6.61GRR194 pKa = 11.84MQPIDD199 pKa = 3.35KK200 pKa = 9.05STSVKK205 pKa = 10.54EE206 pKa = 3.87IDD208 pKa = 3.65STSSGGHH215 pKa = 4.65WNIAGRR221 pKa = 11.84CSCIAYY227 pKa = 8.91HH228 pKa = 6.23QGVALPPITLNRR240 pKa = 11.84HH241 pKa = 5.13NSFCAADD248 pKa = 3.62EE249 pKa = 4.34WFSTHH254 pKa = 6.56PKK256 pKa = 10.39VGTKK260 pKa = 9.93VCISADD266 pKa = 3.36SLAVNFLKK274 pKa = 10.48WSMKK278 pKa = 9.56VMNHH282 pKa = 5.67KK283 pKa = 10.28QDD285 pKa = 3.43IIAVEE290 pKa = 3.97KK291 pKa = 10.66GGTVNYY297 pKa = 10.09IFTKK301 pKa = 9.63RR302 pKa = 11.84AKK304 pKa = 10.1KK305 pKa = 10.58VFISAQMKK313 pKa = 10.09GKK315 pKa = 10.31SCIPGNKK322 pKa = 8.49TGDD325 pKa = 3.5YY326 pKa = 10.4CQEE329 pKa = 4.06KK330 pKa = 11.05VNGCEE335 pKa = 3.92KK336 pKa = 10.96GCWNLPKK343 pKa = 10.49SRR345 pKa = 11.84TRR347 pKa = 11.84QRR349 pKa = 11.84RR350 pKa = 11.84QSILFFWGLVEE361 pKa = 4.38VKK363 pKa = 9.15WGEE366 pKa = 3.97GGEE369 pKa = 4.35VYY371 pKa = 10.69DD372 pKa = 6.09DD373 pKa = 4.17SVLKK377 pKa = 10.86ALASQVDD384 pKa = 4.76LNTASVKK391 pKa = 10.38KK392 pKa = 10.34LQLKK396 pKa = 10.5LDD398 pKa = 4.34LLSKK402 pKa = 10.52AIWNGFCTNGTHH414 pKa = 6.7SVQANMTITFKK425 pKa = 11.2DD426 pKa = 3.19GDD428 pKa = 4.19EE429 pKa = 4.19LVVLDD434 pKa = 4.36WCNSDD439 pKa = 3.35TLEE442 pKa = 4.72AFIDD446 pKa = 3.8SKK448 pKa = 11.16SVQEE452 pKa = 4.18HH453 pKa = 6.28HH454 pKa = 7.19LSDD457 pKa = 3.7LVKK460 pKa = 10.29VSIRR464 pKa = 11.84MRR466 pKa = 11.84IHH468 pKa = 5.68QKK470 pKa = 10.14RR471 pKa = 11.84VTIATKK477 pKa = 10.33LGCVLFGLIVCTAVLRR493 pKa = 11.84ALWRR497 pKa = 11.84RR498 pKa = 11.84KK499 pKa = 10.03DD500 pKa = 3.57FSLSHH505 pKa = 5.13YY506 pKa = 10.6HH507 pKa = 7.17EE508 pKa = 5.38EE509 pKa = 4.24DD510 pKa = 3.97LNGQTCYY517 pKa = 10.18SHH519 pKa = 6.78HH520 pKa = 6.75RR521 pKa = 11.84FEE523 pKa = 4.46KK524 pKa = 10.17RR525 pKa = 11.84YY526 pKa = 10.64SRR528 pKa = 11.84ACRR531 pKa = 11.84CGNVVIGKK539 pKa = 9.69NSFLKK544 pKa = 9.69MAKK547 pKa = 9.87CNNRR551 pKa = 11.84SKK553 pKa = 11.26LPDD556 pKa = 4.3CLLMDD561 pKa = 5.72DD562 pKa = 5.31KK563 pKa = 11.54PNEE566 pKa = 4.12CDD568 pKa = 3.88VLDD571 pKa = 4.58FRR573 pKa = 11.84QGQQKK578 pKa = 9.67EE579 pKa = 4.67VISEE583 pKa = 4.07VQVIEE588 pKa = 4.22FEE590 pKa = 4.13IHH592 pKa = 6.02GDD594 pKa = 3.44EE595 pKa = 4.43PRR597 pKa = 11.84NEE599 pKa = 3.93GSGAQTHH606 pKa = 5.77QDD608 pKa = 3.01LSEE611 pKa = 3.98TPEE614 pKa = 4.03RR615 pKa = 11.84YY616 pKa = 9.66EE617 pKa = 3.55IAPLVFHH624 pKa = 6.48NPEE627 pKa = 4.32YY628 pKa = 10.08VTQFHH633 pKa = 7.06PKK635 pKa = 9.64
MM1 pKa = 7.44FEE3 pKa = 5.2SIIDD7 pKa = 4.85PILEE11 pKa = 3.88WLFNLFSFILAVIFVMTLSLLIYY34 pKa = 10.44RR35 pKa = 11.84MRR37 pKa = 11.84NLPFLFMMCGSAEE50 pKa = 4.03CWVSNIQMNGHH61 pKa = 4.84TAEE64 pKa = 3.87MRR66 pKa = 11.84QYY68 pKa = 10.44EE69 pKa = 4.39GQWLCGKK76 pKa = 10.14IKK78 pKa = 9.88MIKK81 pKa = 10.22LDD83 pKa = 3.57DD84 pKa = 4.23LYY86 pKa = 11.16YY87 pKa = 10.15IRR89 pKa = 11.84YY90 pKa = 9.18NMSDD94 pKa = 3.55SAVHH98 pKa = 6.47LVMSRR103 pKa = 11.84CTNTKK108 pKa = 10.34LEE110 pKa = 4.43NGWEE114 pKa = 4.66SVCSEE119 pKa = 4.34RR120 pKa = 11.84PALLKK125 pKa = 10.85NISTGISMIAGFNCSGQQKK144 pKa = 10.06CFVGDD149 pKa = 3.89GCTPWVDD156 pKa = 3.45ARR158 pKa = 11.84DD159 pKa = 3.3ICGRR163 pKa = 11.84KK164 pKa = 9.56LEE166 pKa = 4.47RR167 pKa = 11.84KK168 pKa = 9.49LDD170 pKa = 3.59NHH172 pKa = 6.56QMLDD176 pKa = 2.92ICRR179 pKa = 11.84QPIVGWIDD187 pKa = 3.17SDD189 pKa = 4.2GVHH192 pKa = 6.61GRR194 pKa = 11.84MQPIDD199 pKa = 3.35KK200 pKa = 9.05STSVKK205 pKa = 10.54EE206 pKa = 3.87IDD208 pKa = 3.65STSSGGHH215 pKa = 4.65WNIAGRR221 pKa = 11.84CSCIAYY227 pKa = 8.91HH228 pKa = 6.23QGVALPPITLNRR240 pKa = 11.84HH241 pKa = 5.13NSFCAADD248 pKa = 3.62EE249 pKa = 4.34WFSTHH254 pKa = 6.56PKK256 pKa = 10.39VGTKK260 pKa = 9.93VCISADD266 pKa = 3.36SLAVNFLKK274 pKa = 10.48WSMKK278 pKa = 9.56VMNHH282 pKa = 5.67KK283 pKa = 10.28QDD285 pKa = 3.43IIAVEE290 pKa = 3.97KK291 pKa = 10.66GGTVNYY297 pKa = 10.09IFTKK301 pKa = 9.63RR302 pKa = 11.84AKK304 pKa = 10.1KK305 pKa = 10.58VFISAQMKK313 pKa = 10.09GKK315 pKa = 10.31SCIPGNKK322 pKa = 8.49TGDD325 pKa = 3.5YY326 pKa = 10.4CQEE329 pKa = 4.06KK330 pKa = 11.05VNGCEE335 pKa = 3.92KK336 pKa = 10.96GCWNLPKK343 pKa = 10.49SRR345 pKa = 11.84TRR347 pKa = 11.84QRR349 pKa = 11.84RR350 pKa = 11.84QSILFFWGLVEE361 pKa = 4.38VKK363 pKa = 9.15WGEE366 pKa = 3.97GGEE369 pKa = 4.35VYY371 pKa = 10.69DD372 pKa = 6.09DD373 pKa = 4.17SVLKK377 pKa = 10.86ALASQVDD384 pKa = 4.76LNTASVKK391 pKa = 10.38KK392 pKa = 10.34LQLKK396 pKa = 10.5LDD398 pKa = 4.34LLSKK402 pKa = 10.52AIWNGFCTNGTHH414 pKa = 6.7SVQANMTITFKK425 pKa = 11.2DD426 pKa = 3.19GDD428 pKa = 4.19EE429 pKa = 4.19LVVLDD434 pKa = 4.36WCNSDD439 pKa = 3.35TLEE442 pKa = 4.72AFIDD446 pKa = 3.8SKK448 pKa = 11.16SVQEE452 pKa = 4.18HH453 pKa = 6.28HH454 pKa = 7.19LSDD457 pKa = 3.7LVKK460 pKa = 10.29VSIRR464 pKa = 11.84MRR466 pKa = 11.84IHH468 pKa = 5.68QKK470 pKa = 10.14RR471 pKa = 11.84VTIATKK477 pKa = 10.33LGCVLFGLIVCTAVLRR493 pKa = 11.84ALWRR497 pKa = 11.84RR498 pKa = 11.84KK499 pKa = 10.03DD500 pKa = 3.57FSLSHH505 pKa = 5.13YY506 pKa = 10.6HH507 pKa = 7.17EE508 pKa = 5.38EE509 pKa = 4.24DD510 pKa = 3.97LNGQTCYY517 pKa = 10.18SHH519 pKa = 6.78HH520 pKa = 6.75RR521 pKa = 11.84FEE523 pKa = 4.46KK524 pKa = 10.17RR525 pKa = 11.84YY526 pKa = 10.64SRR528 pKa = 11.84ACRR531 pKa = 11.84CGNVVIGKK539 pKa = 9.69NSFLKK544 pKa = 9.69MAKK547 pKa = 9.87CNNRR551 pKa = 11.84SKK553 pKa = 11.26LPDD556 pKa = 4.3CLLMDD561 pKa = 5.72DD562 pKa = 5.31KK563 pKa = 11.54PNEE566 pKa = 4.12CDD568 pKa = 3.88VLDD571 pKa = 4.58FRR573 pKa = 11.84QGQQKK578 pKa = 9.67EE579 pKa = 4.67VISEE583 pKa = 4.07VQVIEE588 pKa = 4.22FEE590 pKa = 4.13IHH592 pKa = 6.02GDD594 pKa = 3.44EE595 pKa = 4.43PRR597 pKa = 11.84NEE599 pKa = 3.93GSGAQTHH606 pKa = 5.77QDD608 pKa = 3.01LSEE611 pKa = 3.98TPEE614 pKa = 4.03RR615 pKa = 11.84YY616 pKa = 9.66EE617 pKa = 3.55IAPLVFHH624 pKa = 6.48NPEE627 pKa = 4.32YY628 pKa = 10.08VTQFHH633 pKa = 7.06PKK635 pKa = 9.64
Molecular weight: 72.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3478 |
156 |
2092 |
869.5 |
99.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.112 ± 0.468 | 3.508 ± 0.503 |
5.98 ± 0.848 | 7.418 ± 0.881 |
3.652 ± 0.229 | 5.607 ± 0.399 |
2.53 ± 0.312 | 6.642 ± 0.095 |
7.849 ± 0.483 | 9.804 ± 0.598 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.537 ± 0.408 | 4.255 ± 0.28 |
3.853 ± 0.33 | 2.933 ± 0.419 |
6.009 ± 0.534 | 8.223 ± 0.516 |
4.974 ± 0.362 | 5.463 ± 0.453 |
1.438 ± 0.352 | 2.214 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
