
Rhodobacteraceae bacterium AsT-22
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
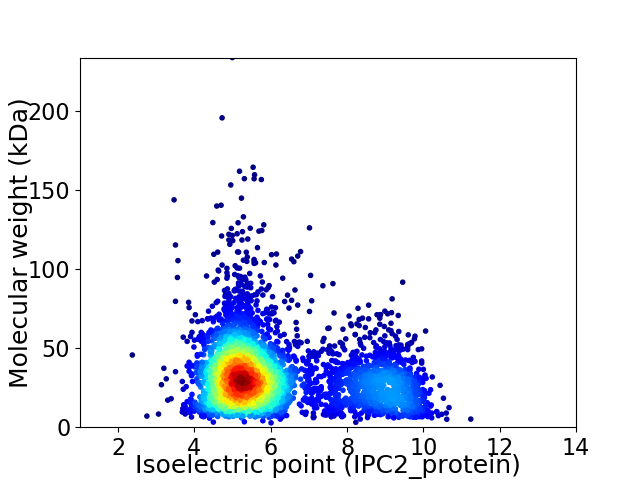
Virtual 2D-PAGE plot for 4150 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A327M1G8|A0A327M1G8_9RHOB Sugar phosphate isomerase/epimerase OS=Rhodobacteraceae bacterium AsT-22 OX=2230886 GN=DOO74_02735 PE=4 SV=1
MM1 pKa = 7.33TSVILTPGFRR11 pKa = 11.84DD12 pKa = 3.6LLEE15 pKa = 5.79DD16 pKa = 3.73DD17 pKa = 4.96TILPGLKK24 pKa = 10.08NFALDD29 pKa = 3.84YY30 pKa = 10.59EE31 pKa = 5.04GEE33 pKa = 4.07LSALGDD39 pKa = 4.04TDD41 pKa = 3.88PAAPPSLSASLIQFFEE57 pKa = 4.06QSTDD61 pKa = 3.03PAYY64 pKa = 9.41PQTYY68 pKa = 8.0TVSFSGSGISPISSIEE84 pKa = 4.02EE85 pKa = 4.07LEE87 pKa = 4.22TALMEE92 pKa = 4.47GLATGTLDD100 pKa = 4.14TVTIDD105 pKa = 3.45YY106 pKa = 10.64GSTEE110 pKa = 3.99ILSLDD115 pKa = 3.69MGSTSYY121 pKa = 10.45TLTTGNQSLEE131 pKa = 4.13FTGAFPDD138 pKa = 3.69TLGDD142 pKa = 4.0FGALVGMASEE152 pKa = 4.63IDD154 pKa = 3.64NIVYY158 pKa = 9.24MSSAEE163 pKa = 4.02RR164 pKa = 11.84EE165 pKa = 4.24AFLAPLMEE173 pKa = 4.08YY174 pKa = 10.44DD175 pKa = 2.96ITEE178 pKa = 4.0VVLRR182 pKa = 11.84DD183 pKa = 3.48SGTEE187 pKa = 3.9LLSLGVDD194 pKa = 4.97FATGSYY200 pKa = 8.67TVAAGGYY207 pKa = 7.48TLDD210 pKa = 4.05ATITTPPLHH219 pKa = 6.62EE220 pKa = 5.07LLNTLLQMEE229 pKa = 4.7MEE231 pKa = 4.05WGEE234 pKa = 4.23GGVQSPHH241 pKa = 5.95LRR243 pKa = 11.84LYY245 pKa = 10.27DD246 pKa = 3.6ASGTLVAEE254 pKa = 4.62NADD257 pKa = 3.76TSDD260 pKa = 3.36PGSPHH265 pKa = 5.49FHH267 pKa = 6.43GGYY270 pKa = 10.41AYY272 pKa = 10.57FSYY275 pKa = 10.47TPTVSEE281 pKa = 3.97TFYY284 pKa = 10.77MFGASVGDD292 pKa = 3.89AGIGFYY298 pKa = 11.44DD299 pKa = 3.22MGFWMSGSTGDD310 pKa = 3.31WTEE313 pKa = 3.81LSEE316 pKa = 5.66DD317 pKa = 3.48ADD319 pKa = 4.0APADD323 pKa = 3.46ATTPYY328 pKa = 10.11IFNGPGTFTFNGVLFPEE345 pKa = 4.95ADD347 pKa = 4.12RR348 pKa = 11.84DD349 pKa = 3.75WVAVEE354 pKa = 4.87LEE356 pKa = 4.52ADD358 pKa = 3.61TEE360 pKa = 4.75YY361 pKa = 10.94QFSMSGFFEE370 pKa = 4.66PPWEE374 pKa = 4.17FEE376 pKa = 4.37GYY378 pKa = 7.98TLGPITLTDD387 pKa = 3.69PMGEE391 pKa = 4.39TILHH395 pKa = 5.91IPEE398 pKa = 4.49TDD400 pKa = 3.27LTDD403 pKa = 4.32AMALQALIDD412 pKa = 4.7EE413 pKa = 4.57ISILLEE419 pKa = 4.16GLGLPPLPEE428 pKa = 4.67GLLGLNNGDD437 pKa = 3.64PHH439 pKa = 8.61LLTLDD444 pKa = 3.56GAAYY448 pKa = 9.87DD449 pKa = 3.77FHH451 pKa = 8.27AAGEE455 pKa = 4.41YY456 pKa = 10.27VLTRR460 pKa = 11.84ATDD463 pKa = 3.69GSDD466 pKa = 3.71FEE468 pKa = 4.64VQARR472 pKa = 11.84MSPVGEE478 pKa = 3.76NVTANVAAGVRR489 pKa = 11.84LDD491 pKa = 3.95GGNVMVDD498 pKa = 3.4AAAANPLTVDD508 pKa = 4.08GVATAVADD516 pKa = 3.77GGFILVGQDD525 pKa = 2.41RR526 pKa = 11.84VYY528 pKa = 11.08RR529 pKa = 11.84EE530 pKa = 3.7GDD532 pKa = 3.64TYY534 pKa = 11.19TLIHH538 pKa = 6.29TRR540 pKa = 11.84DD541 pKa = 3.4GDD543 pKa = 3.97LEE545 pKa = 4.32TGYY548 pKa = 10.7SAVVVGVVGGRR559 pKa = 11.84VDD561 pKa = 2.99ITVALDD567 pKa = 4.2GYY569 pKa = 10.03WGGNVEE575 pKa = 4.38GLLGNADD582 pKa = 3.42GNAANDD588 pKa = 3.15IALADD593 pKa = 4.04GTPLDD598 pKa = 4.58RR599 pKa = 11.84PLKK602 pKa = 10.28FDD604 pKa = 4.33DD605 pKa = 4.83VYY607 pKa = 11.5GQYY610 pKa = 10.36RR611 pKa = 11.84DD612 pKa = 3.24DD613 pKa = 3.66WRR615 pKa = 11.84VDD617 pKa = 3.46DD618 pKa = 5.2AADD621 pKa = 3.49SLFSYY626 pKa = 10.51GAGEE630 pKa = 4.78GPDD633 pKa = 3.95SYY635 pKa = 11.71YY636 pKa = 10.98LPNYY640 pKa = 5.52PTGMIGLDD648 pKa = 3.43NFDD651 pKa = 4.5PADD654 pKa = 3.44VSAAEE659 pKa = 4.18AVVTAGGLAPGTLAFQQAVLDD680 pKa = 3.95YY681 pKa = 10.97LLTEE685 pKa = 4.52DD686 pKa = 4.51EE687 pKa = 5.01SYY689 pKa = 11.27IDD691 pKa = 3.64TATNTQTAIDD701 pKa = 4.18SRR703 pKa = 11.84PAEE706 pKa = 4.28APAIEE711 pKa = 4.54TPDD714 pKa = 3.35TDD716 pKa = 3.59GGGLEE721 pKa = 4.33GLLTLSGKK729 pKa = 8.27LTSLAGEE736 pKa = 5.26DD737 pKa = 3.14ITGATVTFQPTGRR750 pKa = 11.84SVSLARR756 pKa = 11.84LTRR759 pKa = 11.84DD760 pKa = 3.53DD761 pKa = 4.51GDD763 pKa = 3.72FSFDD767 pKa = 3.29MVAGEE772 pKa = 4.82DD773 pKa = 3.35GHH775 pKa = 8.59LNATRR780 pKa = 11.84GYY782 pKa = 10.87DD783 pKa = 3.32ADD785 pKa = 3.99TDD787 pKa = 4.1PGINAGDD794 pKa = 3.87ALDD797 pKa = 3.69VLRR800 pKa = 11.84IAVGLPPSFGPAEE813 pKa = 4.13AQNFVAADD821 pKa = 3.22IDD823 pKa = 3.84GDD825 pKa = 3.73RR826 pKa = 11.84RR827 pKa = 11.84ATAGDD832 pKa = 3.51ALDD835 pKa = 3.91VLRR838 pKa = 11.84HH839 pKa = 4.88AVGLEE844 pKa = 4.26SEE846 pKa = 4.87HH847 pKa = 5.97TPHH850 pKa = 5.91WTFFAADD857 pKa = 4.09TDD859 pKa = 3.82WDD861 pKa = 4.11ALDD864 pKa = 5.15LGASNTSVSSGAAVDD879 pKa = 4.2ALAANFDD886 pKa = 3.82VPMTGILIGNMEE898 pKa = 4.41TVVGG902 pKa = 4.05
MM1 pKa = 7.33TSVILTPGFRR11 pKa = 11.84DD12 pKa = 3.6LLEE15 pKa = 5.79DD16 pKa = 3.73DD17 pKa = 4.96TILPGLKK24 pKa = 10.08NFALDD29 pKa = 3.84YY30 pKa = 10.59EE31 pKa = 5.04GEE33 pKa = 4.07LSALGDD39 pKa = 4.04TDD41 pKa = 3.88PAAPPSLSASLIQFFEE57 pKa = 4.06QSTDD61 pKa = 3.03PAYY64 pKa = 9.41PQTYY68 pKa = 8.0TVSFSGSGISPISSIEE84 pKa = 4.02EE85 pKa = 4.07LEE87 pKa = 4.22TALMEE92 pKa = 4.47GLATGTLDD100 pKa = 4.14TVTIDD105 pKa = 3.45YY106 pKa = 10.64GSTEE110 pKa = 3.99ILSLDD115 pKa = 3.69MGSTSYY121 pKa = 10.45TLTTGNQSLEE131 pKa = 4.13FTGAFPDD138 pKa = 3.69TLGDD142 pKa = 4.0FGALVGMASEE152 pKa = 4.63IDD154 pKa = 3.64NIVYY158 pKa = 9.24MSSAEE163 pKa = 4.02RR164 pKa = 11.84EE165 pKa = 4.24AFLAPLMEE173 pKa = 4.08YY174 pKa = 10.44DD175 pKa = 2.96ITEE178 pKa = 4.0VVLRR182 pKa = 11.84DD183 pKa = 3.48SGTEE187 pKa = 3.9LLSLGVDD194 pKa = 4.97FATGSYY200 pKa = 8.67TVAAGGYY207 pKa = 7.48TLDD210 pKa = 4.05ATITTPPLHH219 pKa = 6.62EE220 pKa = 5.07LLNTLLQMEE229 pKa = 4.7MEE231 pKa = 4.05WGEE234 pKa = 4.23GGVQSPHH241 pKa = 5.95LRR243 pKa = 11.84LYY245 pKa = 10.27DD246 pKa = 3.6ASGTLVAEE254 pKa = 4.62NADD257 pKa = 3.76TSDD260 pKa = 3.36PGSPHH265 pKa = 5.49FHH267 pKa = 6.43GGYY270 pKa = 10.41AYY272 pKa = 10.57FSYY275 pKa = 10.47TPTVSEE281 pKa = 3.97TFYY284 pKa = 10.77MFGASVGDD292 pKa = 3.89AGIGFYY298 pKa = 11.44DD299 pKa = 3.22MGFWMSGSTGDD310 pKa = 3.31WTEE313 pKa = 3.81LSEE316 pKa = 5.66DD317 pKa = 3.48ADD319 pKa = 4.0APADD323 pKa = 3.46ATTPYY328 pKa = 10.11IFNGPGTFTFNGVLFPEE345 pKa = 4.95ADD347 pKa = 4.12RR348 pKa = 11.84DD349 pKa = 3.75WVAVEE354 pKa = 4.87LEE356 pKa = 4.52ADD358 pKa = 3.61TEE360 pKa = 4.75YY361 pKa = 10.94QFSMSGFFEE370 pKa = 4.66PPWEE374 pKa = 4.17FEE376 pKa = 4.37GYY378 pKa = 7.98TLGPITLTDD387 pKa = 3.69PMGEE391 pKa = 4.39TILHH395 pKa = 5.91IPEE398 pKa = 4.49TDD400 pKa = 3.27LTDD403 pKa = 4.32AMALQALIDD412 pKa = 4.7EE413 pKa = 4.57ISILLEE419 pKa = 4.16GLGLPPLPEE428 pKa = 4.67GLLGLNNGDD437 pKa = 3.64PHH439 pKa = 8.61LLTLDD444 pKa = 3.56GAAYY448 pKa = 9.87DD449 pKa = 3.77FHH451 pKa = 8.27AAGEE455 pKa = 4.41YY456 pKa = 10.27VLTRR460 pKa = 11.84ATDD463 pKa = 3.69GSDD466 pKa = 3.71FEE468 pKa = 4.64VQARR472 pKa = 11.84MSPVGEE478 pKa = 3.76NVTANVAAGVRR489 pKa = 11.84LDD491 pKa = 3.95GGNVMVDD498 pKa = 3.4AAAANPLTVDD508 pKa = 4.08GVATAVADD516 pKa = 3.77GGFILVGQDD525 pKa = 2.41RR526 pKa = 11.84VYY528 pKa = 11.08RR529 pKa = 11.84EE530 pKa = 3.7GDD532 pKa = 3.64TYY534 pKa = 11.19TLIHH538 pKa = 6.29TRR540 pKa = 11.84DD541 pKa = 3.4GDD543 pKa = 3.97LEE545 pKa = 4.32TGYY548 pKa = 10.7SAVVVGVVGGRR559 pKa = 11.84VDD561 pKa = 2.99ITVALDD567 pKa = 4.2GYY569 pKa = 10.03WGGNVEE575 pKa = 4.38GLLGNADD582 pKa = 3.42GNAANDD588 pKa = 3.15IALADD593 pKa = 4.04GTPLDD598 pKa = 4.58RR599 pKa = 11.84PLKK602 pKa = 10.28FDD604 pKa = 4.33DD605 pKa = 4.83VYY607 pKa = 11.5GQYY610 pKa = 10.36RR611 pKa = 11.84DD612 pKa = 3.24DD613 pKa = 3.66WRR615 pKa = 11.84VDD617 pKa = 3.46DD618 pKa = 5.2AADD621 pKa = 3.49SLFSYY626 pKa = 10.51GAGEE630 pKa = 4.78GPDD633 pKa = 3.95SYY635 pKa = 11.71YY636 pKa = 10.98LPNYY640 pKa = 5.52PTGMIGLDD648 pKa = 3.43NFDD651 pKa = 4.5PADD654 pKa = 3.44VSAAEE659 pKa = 4.18AVVTAGGLAPGTLAFQQAVLDD680 pKa = 3.95YY681 pKa = 10.97LLTEE685 pKa = 4.52DD686 pKa = 4.51EE687 pKa = 5.01SYY689 pKa = 11.27IDD691 pKa = 3.64TATNTQTAIDD701 pKa = 4.18SRR703 pKa = 11.84PAEE706 pKa = 4.28APAIEE711 pKa = 4.54TPDD714 pKa = 3.35TDD716 pKa = 3.59GGGLEE721 pKa = 4.33GLLTLSGKK729 pKa = 8.27LTSLAGEE736 pKa = 5.26DD737 pKa = 3.14ITGATVTFQPTGRR750 pKa = 11.84SVSLARR756 pKa = 11.84LTRR759 pKa = 11.84DD760 pKa = 3.53DD761 pKa = 4.51GDD763 pKa = 3.72FSFDD767 pKa = 3.29MVAGEE772 pKa = 4.82DD773 pKa = 3.35GHH775 pKa = 8.59LNATRR780 pKa = 11.84GYY782 pKa = 10.87DD783 pKa = 3.32ADD785 pKa = 3.99TDD787 pKa = 4.1PGINAGDD794 pKa = 3.87ALDD797 pKa = 3.69VLRR800 pKa = 11.84IAVGLPPSFGPAEE813 pKa = 4.13AQNFVAADD821 pKa = 3.22IDD823 pKa = 3.84GDD825 pKa = 3.73RR826 pKa = 11.84RR827 pKa = 11.84ATAGDD832 pKa = 3.51ALDD835 pKa = 3.91VLRR838 pKa = 11.84HH839 pKa = 4.88AVGLEE844 pKa = 4.26SEE846 pKa = 4.87HH847 pKa = 5.97TPHH850 pKa = 5.91WTFFAADD857 pKa = 4.09TDD859 pKa = 3.82WDD861 pKa = 4.11ALDD864 pKa = 5.15LGASNTSVSSGAAVDD879 pKa = 4.2ALAANFDD886 pKa = 3.82VPMTGILIGNMEE898 pKa = 4.41TVVGG902 pKa = 4.05
Molecular weight: 94.79 kDa
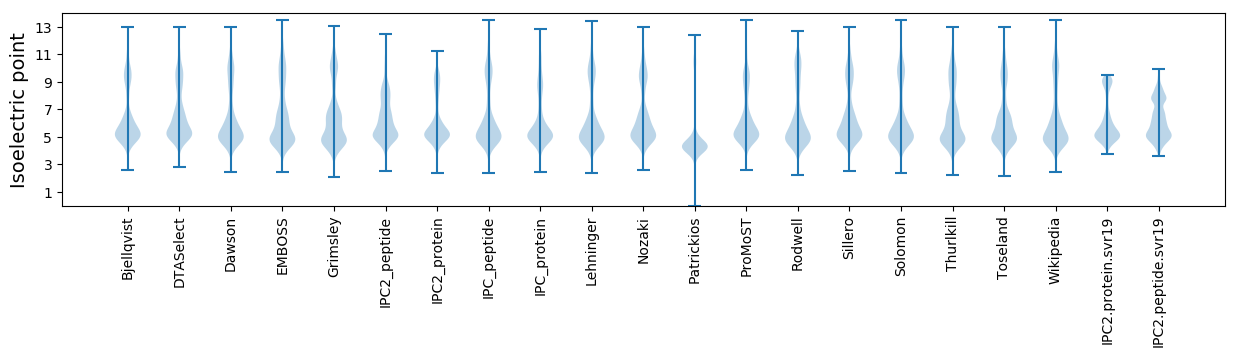
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A327LRW8|A0A327LRW8_9RHOB Uncharacterized protein OS=Rhodobacteraceae bacterium AsT-22 OX=2230886 GN=DOO74_13700 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1294212 |
26 |
2150 |
311.9 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.233 ± 0.043 | 0.888 ± 0.011 |
6.093 ± 0.027 | 6.1 ± 0.039 |
3.712 ± 0.023 | 8.904 ± 0.034 |
2.172 ± 0.02 | 5.212 ± 0.028 |
2.929 ± 0.03 | 9.977 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.894 ± 0.017 | 2.478 ± 0.016 |
5.199 ± 0.031 | 2.987 ± 0.021 |
7.151 ± 0.039 | 4.92 ± 0.025 |
5.304 ± 0.024 | 7.28 ± 0.032 |
1.408 ± 0.017 | 2.162 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
