
Shahe narna-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.62
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KIK7|A0A1L3KIK7_9VIRU RNA-dependent RNA polymerase OS=Shahe narna-like virus 7 OX=1923435 PE=4 SV=1
MM1 pKa = 7.68PSWLFTSHH9 pKa = 5.88EE10 pKa = 4.32SVRR13 pKa = 11.84IGMTVLTISSGFNLPVSVNLDD34 pKa = 3.81PIMTPSKK41 pKa = 10.39SIPSSMEE48 pKa = 3.11SWEE51 pKa = 3.82IDD53 pKa = 3.63LVCRR57 pKa = 11.84FLGIKK62 pKa = 9.68EE63 pKa = 4.62EE64 pKa = 4.14YY65 pKa = 10.36VEE67 pKa = 3.91WFMFHH72 pKa = 7.14HH73 pKa = 5.95STKK76 pKa = 10.41KK77 pKa = 10.67GPNGQALMSSTHH89 pKa = 7.21DD90 pKa = 3.65FTLMPSWLKK99 pKa = 10.84NKK101 pKa = 9.77LMSLAGVGLKK111 pKa = 9.85QCFDD115 pKa = 5.1LIDD118 pKa = 4.04SVSMNGISVNSWWNRR133 pKa = 11.84IYY135 pKa = 10.32PVRR138 pKa = 11.84SSTISKK144 pKa = 8.41LTGMSDD150 pKa = 3.45KK151 pKa = 10.8EE152 pKa = 4.46GKK154 pKa = 7.96TSVMGVVDD162 pKa = 3.97YY163 pKa = 10.35WSQTALKK170 pKa = 8.66PLHH173 pKa = 6.41DD174 pKa = 3.71TLMRR178 pKa = 11.84ILSGIGPDD186 pKa = 3.79CTHH189 pKa = 7.09DD190 pKa = 3.44QQSFLTKK197 pKa = 9.89VPSSGVTFYY206 pKa = 11.21SYY208 pKa = 11.81DD209 pKa = 3.41LTNATDD215 pKa = 5.34SMPLWLQGEE224 pKa = 4.61IISRR228 pKa = 11.84MIGKK232 pKa = 9.22DD233 pKa = 3.11RR234 pKa = 11.84AEE236 pKa = 4.01DD237 pKa = 3.29WKK239 pKa = 11.19DD240 pKa = 3.11ILVKK244 pKa = 10.93EE245 pKa = 4.14EE246 pKa = 4.46FSLKK250 pKa = 10.1GHH252 pKa = 5.72PAKK255 pKa = 10.47IKK257 pKa = 10.37FSAGQPMGAYY267 pKa = 10.15SSWPSMALTHH277 pKa = 6.21HH278 pKa = 7.25AIVQLAWISLGNKK291 pKa = 9.66SPTRR295 pKa = 11.84DD296 pKa = 3.54YY297 pKa = 11.85VLLGDD302 pKa = 6.2DD303 pKa = 3.49MVIWNDD309 pKa = 3.38KK310 pKa = 10.56LAMSYY315 pKa = 8.44RR316 pKa = 11.84TLLSSLDD323 pKa = 3.77MPISDD328 pKa = 3.73QKK330 pKa = 8.95THH332 pKa = 6.42ISGSTFEE339 pKa = 4.73FCKK342 pKa = 10.24SWFVNGVEE350 pKa = 4.24VTGFSVPGLWEE361 pKa = 3.99VSSSYY366 pKa = 11.51SKK368 pKa = 10.75LANFLEE374 pKa = 4.55NQEE377 pKa = 4.1SHH379 pKa = 5.15GWKK382 pKa = 10.02PSDD385 pKa = 3.55ASAIPLIMSLYY396 pKa = 9.69NCLGLHH402 pKa = 5.43SQGKK406 pKa = 8.84SVYY409 pKa = 10.1KK410 pKa = 10.51LYY412 pKa = 10.4TVFQTLVKK420 pKa = 9.68TKK422 pKa = 9.52VTGEE426 pKa = 3.99YY427 pKa = 9.57STMAKK432 pKa = 10.65AMTEE436 pKa = 3.82AFRR439 pKa = 11.84WPFYY443 pKa = 8.21EE444 pKa = 3.93TSNEE448 pKa = 3.54IFASILGKK456 pKa = 9.84MKK458 pKa = 10.52EE459 pKa = 4.05EE460 pKa = 3.98QVWKK464 pKa = 10.71DD465 pKa = 3.45MEE467 pKa = 4.41SLQTNVYY474 pKa = 10.02LFHH477 pKa = 8.04DD478 pKa = 4.51EE479 pKa = 4.11FNKK482 pKa = 10.33RR483 pKa = 11.84MSTVNPLWASEE494 pKa = 4.4SYY496 pKa = 8.59KK497 pKa = 9.71TAYY500 pKa = 8.48TEE502 pKa = 3.95RR503 pKa = 11.84VPMLIMAEE511 pKa = 4.13YY512 pKa = 8.96LTMEE516 pKa = 4.58LEE518 pKa = 4.35EE519 pKa = 5.01KK520 pKa = 10.99LMILTGDD527 pKa = 3.66NSSDD531 pKa = 3.45EE532 pKa = 4.12YY533 pKa = 11.42QEE535 pKa = 5.79LIFNEE540 pKa = 4.23SLGSYY545 pKa = 9.84KK546 pKa = 10.4LSKK549 pKa = 10.65HH550 pKa = 5.26FFGLKK555 pKa = 10.35DD556 pKa = 3.41EE557 pKa = 4.83ASSVRR562 pKa = 11.84DD563 pKa = 3.14LSRR566 pKa = 11.84LVKK569 pKa = 10.1PLINKK574 pKa = 9.2VSEE577 pKa = 4.8IINKK581 pKa = 9.58SDD583 pKa = 3.13PTTSVILTTTEE594 pKa = 3.58QMFVKK599 pKa = 9.87DD600 pKa = 3.54TATFGPP606 pKa = 4.18
MM1 pKa = 7.68PSWLFTSHH9 pKa = 5.88EE10 pKa = 4.32SVRR13 pKa = 11.84IGMTVLTISSGFNLPVSVNLDD34 pKa = 3.81PIMTPSKK41 pKa = 10.39SIPSSMEE48 pKa = 3.11SWEE51 pKa = 3.82IDD53 pKa = 3.63LVCRR57 pKa = 11.84FLGIKK62 pKa = 9.68EE63 pKa = 4.62EE64 pKa = 4.14YY65 pKa = 10.36VEE67 pKa = 3.91WFMFHH72 pKa = 7.14HH73 pKa = 5.95STKK76 pKa = 10.41KK77 pKa = 10.67GPNGQALMSSTHH89 pKa = 7.21DD90 pKa = 3.65FTLMPSWLKK99 pKa = 10.84NKK101 pKa = 9.77LMSLAGVGLKK111 pKa = 9.85QCFDD115 pKa = 5.1LIDD118 pKa = 4.04SVSMNGISVNSWWNRR133 pKa = 11.84IYY135 pKa = 10.32PVRR138 pKa = 11.84SSTISKK144 pKa = 8.41LTGMSDD150 pKa = 3.45KK151 pKa = 10.8EE152 pKa = 4.46GKK154 pKa = 7.96TSVMGVVDD162 pKa = 3.97YY163 pKa = 10.35WSQTALKK170 pKa = 8.66PLHH173 pKa = 6.41DD174 pKa = 3.71TLMRR178 pKa = 11.84ILSGIGPDD186 pKa = 3.79CTHH189 pKa = 7.09DD190 pKa = 3.44QQSFLTKK197 pKa = 9.89VPSSGVTFYY206 pKa = 11.21SYY208 pKa = 11.81DD209 pKa = 3.41LTNATDD215 pKa = 5.34SMPLWLQGEE224 pKa = 4.61IISRR228 pKa = 11.84MIGKK232 pKa = 9.22DD233 pKa = 3.11RR234 pKa = 11.84AEE236 pKa = 4.01DD237 pKa = 3.29WKK239 pKa = 11.19DD240 pKa = 3.11ILVKK244 pKa = 10.93EE245 pKa = 4.14EE246 pKa = 4.46FSLKK250 pKa = 10.1GHH252 pKa = 5.72PAKK255 pKa = 10.47IKK257 pKa = 10.37FSAGQPMGAYY267 pKa = 10.15SSWPSMALTHH277 pKa = 6.21HH278 pKa = 7.25AIVQLAWISLGNKK291 pKa = 9.66SPTRR295 pKa = 11.84DD296 pKa = 3.54YY297 pKa = 11.85VLLGDD302 pKa = 6.2DD303 pKa = 3.49MVIWNDD309 pKa = 3.38KK310 pKa = 10.56LAMSYY315 pKa = 8.44RR316 pKa = 11.84TLLSSLDD323 pKa = 3.77MPISDD328 pKa = 3.73QKK330 pKa = 8.95THH332 pKa = 6.42ISGSTFEE339 pKa = 4.73FCKK342 pKa = 10.24SWFVNGVEE350 pKa = 4.24VTGFSVPGLWEE361 pKa = 3.99VSSSYY366 pKa = 11.51SKK368 pKa = 10.75LANFLEE374 pKa = 4.55NQEE377 pKa = 4.1SHH379 pKa = 5.15GWKK382 pKa = 10.02PSDD385 pKa = 3.55ASAIPLIMSLYY396 pKa = 9.69NCLGLHH402 pKa = 5.43SQGKK406 pKa = 8.84SVYY409 pKa = 10.1KK410 pKa = 10.51LYY412 pKa = 10.4TVFQTLVKK420 pKa = 9.68TKK422 pKa = 9.52VTGEE426 pKa = 3.99YY427 pKa = 9.57STMAKK432 pKa = 10.65AMTEE436 pKa = 3.82AFRR439 pKa = 11.84WPFYY443 pKa = 8.21EE444 pKa = 3.93TSNEE448 pKa = 3.54IFASILGKK456 pKa = 9.84MKK458 pKa = 10.52EE459 pKa = 4.05EE460 pKa = 3.98QVWKK464 pKa = 10.71DD465 pKa = 3.45MEE467 pKa = 4.41SLQTNVYY474 pKa = 10.02LFHH477 pKa = 8.04DD478 pKa = 4.51EE479 pKa = 4.11FNKK482 pKa = 10.33RR483 pKa = 11.84MSTVNPLWASEE494 pKa = 4.4SYY496 pKa = 8.59KK497 pKa = 9.71TAYY500 pKa = 8.48TEE502 pKa = 3.95RR503 pKa = 11.84VPMLIMAEE511 pKa = 4.13YY512 pKa = 8.96LTMEE516 pKa = 4.58LEE518 pKa = 4.35EE519 pKa = 5.01KK520 pKa = 10.99LMILTGDD527 pKa = 3.66NSSDD531 pKa = 3.45EE532 pKa = 4.12YY533 pKa = 11.42QEE535 pKa = 5.79LIFNEE540 pKa = 4.23SLGSYY545 pKa = 9.84KK546 pKa = 10.4LSKK549 pKa = 10.65HH550 pKa = 5.26FFGLKK555 pKa = 10.35DD556 pKa = 3.41EE557 pKa = 4.83ASSVRR562 pKa = 11.84DD563 pKa = 3.14LSRR566 pKa = 11.84LVKK569 pKa = 10.1PLINKK574 pKa = 9.2VSEE577 pKa = 4.8IINKK581 pKa = 9.58SDD583 pKa = 3.13PTTSVILTTTEE594 pKa = 3.58QMFVKK599 pKa = 9.87DD600 pKa = 3.54TATFGPP606 pKa = 4.18
Molecular weight: 68.42 kDa
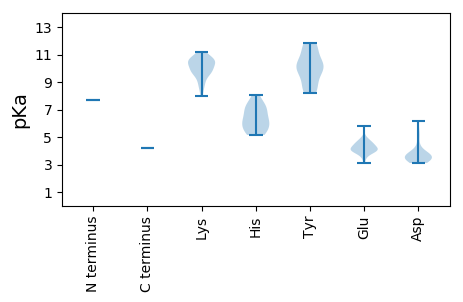
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIK7|A0A1L3KIK7_9VIRU RNA-dependent RNA polymerase OS=Shahe narna-like virus 7 OX=1923435 PE=4 SV=1
MM1 pKa = 7.68PSWLFTSHH9 pKa = 5.88EE10 pKa = 4.32SVRR13 pKa = 11.84IGMTVLTISSGFNLPVSVNLDD34 pKa = 3.81PIMTPSKK41 pKa = 10.39SIPSSMEE48 pKa = 3.11SWEE51 pKa = 3.82IDD53 pKa = 3.63LVCRR57 pKa = 11.84FLGIKK62 pKa = 9.68EE63 pKa = 4.62EE64 pKa = 4.14YY65 pKa = 10.36VEE67 pKa = 3.91WFMFHH72 pKa = 7.14HH73 pKa = 5.95STKK76 pKa = 10.41KK77 pKa = 10.67GPNGQALMSSTHH89 pKa = 7.21DD90 pKa = 3.65FTLMPSWLKK99 pKa = 10.84NKK101 pKa = 9.77LMSLAGVGLKK111 pKa = 9.85QCFDD115 pKa = 5.1LIDD118 pKa = 4.04SVSMNGISVNSWWNRR133 pKa = 11.84IYY135 pKa = 10.32PVRR138 pKa = 11.84SSTISKK144 pKa = 8.41LTGMSDD150 pKa = 3.45KK151 pKa = 10.8EE152 pKa = 4.46GKK154 pKa = 7.96TSVMGVVDD162 pKa = 3.97YY163 pKa = 10.35WSQTALKK170 pKa = 8.66PLHH173 pKa = 6.41DD174 pKa = 3.71TLMRR178 pKa = 11.84ILSGIGPDD186 pKa = 3.79CTHH189 pKa = 7.09DD190 pKa = 3.44QQSFLTKK197 pKa = 9.89VPSSGVTFYY206 pKa = 11.21SYY208 pKa = 11.81DD209 pKa = 3.41LTNATDD215 pKa = 5.34SMPLWLQGEE224 pKa = 4.61IISRR228 pKa = 11.84MIGKK232 pKa = 9.22DD233 pKa = 3.11RR234 pKa = 11.84AEE236 pKa = 4.01DD237 pKa = 3.29WKK239 pKa = 11.19DD240 pKa = 3.11ILVKK244 pKa = 10.93EE245 pKa = 4.14EE246 pKa = 4.46FSLKK250 pKa = 10.1GHH252 pKa = 5.72PAKK255 pKa = 10.47IKK257 pKa = 10.37FSAGQPMGAYY267 pKa = 10.15SSWPSMALTHH277 pKa = 6.21HH278 pKa = 7.25AIVQLAWISLGNKK291 pKa = 9.66SPTRR295 pKa = 11.84DD296 pKa = 3.54YY297 pKa = 11.85VLLGDD302 pKa = 6.2DD303 pKa = 3.49MVIWNDD309 pKa = 3.38KK310 pKa = 10.56LAMSYY315 pKa = 8.44RR316 pKa = 11.84TLLSSLDD323 pKa = 3.77MPISDD328 pKa = 3.73QKK330 pKa = 8.95THH332 pKa = 6.42ISGSTFEE339 pKa = 4.73FCKK342 pKa = 10.24SWFVNGVEE350 pKa = 4.24VTGFSVPGLWEE361 pKa = 3.99VSSSYY366 pKa = 11.51SKK368 pKa = 10.75LANFLEE374 pKa = 4.55NQEE377 pKa = 4.1SHH379 pKa = 5.15GWKK382 pKa = 10.02PSDD385 pKa = 3.55ASAIPLIMSLYY396 pKa = 9.69NCLGLHH402 pKa = 5.43SQGKK406 pKa = 8.84SVYY409 pKa = 10.1KK410 pKa = 10.51LYY412 pKa = 10.4TVFQTLVKK420 pKa = 9.68TKK422 pKa = 9.52VTGEE426 pKa = 3.99YY427 pKa = 9.57STMAKK432 pKa = 10.65AMTEE436 pKa = 3.82AFRR439 pKa = 11.84WPFYY443 pKa = 8.21EE444 pKa = 3.93TSNEE448 pKa = 3.54IFASILGKK456 pKa = 9.84MKK458 pKa = 10.52EE459 pKa = 4.05EE460 pKa = 3.98QVWKK464 pKa = 10.71DD465 pKa = 3.45MEE467 pKa = 4.41SLQTNVYY474 pKa = 10.02LFHH477 pKa = 8.04DD478 pKa = 4.51EE479 pKa = 4.11FNKK482 pKa = 10.33RR483 pKa = 11.84MSTVNPLWASEE494 pKa = 4.4SYY496 pKa = 8.59KK497 pKa = 9.71TAYY500 pKa = 8.48TEE502 pKa = 3.95RR503 pKa = 11.84VPMLIMAEE511 pKa = 4.13YY512 pKa = 8.96LTMEE516 pKa = 4.58LEE518 pKa = 4.35EE519 pKa = 5.01KK520 pKa = 10.99LMILTGDD527 pKa = 3.66NSSDD531 pKa = 3.45EE532 pKa = 4.12YY533 pKa = 11.42QEE535 pKa = 5.79LIFNEE540 pKa = 4.23SLGSYY545 pKa = 9.84KK546 pKa = 10.4LSKK549 pKa = 10.65HH550 pKa = 5.26FFGLKK555 pKa = 10.35DD556 pKa = 3.41EE557 pKa = 4.83ASSVRR562 pKa = 11.84DD563 pKa = 3.14LSRR566 pKa = 11.84LVKK569 pKa = 10.1PLINKK574 pKa = 9.2VSEE577 pKa = 4.8IINKK581 pKa = 9.58SDD583 pKa = 3.13PTTSVILTTTEE594 pKa = 3.58QMFVKK599 pKa = 9.87DD600 pKa = 3.54TATFGPP606 pKa = 4.18
MM1 pKa = 7.68PSWLFTSHH9 pKa = 5.88EE10 pKa = 4.32SVRR13 pKa = 11.84IGMTVLTISSGFNLPVSVNLDD34 pKa = 3.81PIMTPSKK41 pKa = 10.39SIPSSMEE48 pKa = 3.11SWEE51 pKa = 3.82IDD53 pKa = 3.63LVCRR57 pKa = 11.84FLGIKK62 pKa = 9.68EE63 pKa = 4.62EE64 pKa = 4.14YY65 pKa = 10.36VEE67 pKa = 3.91WFMFHH72 pKa = 7.14HH73 pKa = 5.95STKK76 pKa = 10.41KK77 pKa = 10.67GPNGQALMSSTHH89 pKa = 7.21DD90 pKa = 3.65FTLMPSWLKK99 pKa = 10.84NKK101 pKa = 9.77LMSLAGVGLKK111 pKa = 9.85QCFDD115 pKa = 5.1LIDD118 pKa = 4.04SVSMNGISVNSWWNRR133 pKa = 11.84IYY135 pKa = 10.32PVRR138 pKa = 11.84SSTISKK144 pKa = 8.41LTGMSDD150 pKa = 3.45KK151 pKa = 10.8EE152 pKa = 4.46GKK154 pKa = 7.96TSVMGVVDD162 pKa = 3.97YY163 pKa = 10.35WSQTALKK170 pKa = 8.66PLHH173 pKa = 6.41DD174 pKa = 3.71TLMRR178 pKa = 11.84ILSGIGPDD186 pKa = 3.79CTHH189 pKa = 7.09DD190 pKa = 3.44QQSFLTKK197 pKa = 9.89VPSSGVTFYY206 pKa = 11.21SYY208 pKa = 11.81DD209 pKa = 3.41LTNATDD215 pKa = 5.34SMPLWLQGEE224 pKa = 4.61IISRR228 pKa = 11.84MIGKK232 pKa = 9.22DD233 pKa = 3.11RR234 pKa = 11.84AEE236 pKa = 4.01DD237 pKa = 3.29WKK239 pKa = 11.19DD240 pKa = 3.11ILVKK244 pKa = 10.93EE245 pKa = 4.14EE246 pKa = 4.46FSLKK250 pKa = 10.1GHH252 pKa = 5.72PAKK255 pKa = 10.47IKK257 pKa = 10.37FSAGQPMGAYY267 pKa = 10.15SSWPSMALTHH277 pKa = 6.21HH278 pKa = 7.25AIVQLAWISLGNKK291 pKa = 9.66SPTRR295 pKa = 11.84DD296 pKa = 3.54YY297 pKa = 11.85VLLGDD302 pKa = 6.2DD303 pKa = 3.49MVIWNDD309 pKa = 3.38KK310 pKa = 10.56LAMSYY315 pKa = 8.44RR316 pKa = 11.84TLLSSLDD323 pKa = 3.77MPISDD328 pKa = 3.73QKK330 pKa = 8.95THH332 pKa = 6.42ISGSTFEE339 pKa = 4.73FCKK342 pKa = 10.24SWFVNGVEE350 pKa = 4.24VTGFSVPGLWEE361 pKa = 3.99VSSSYY366 pKa = 11.51SKK368 pKa = 10.75LANFLEE374 pKa = 4.55NQEE377 pKa = 4.1SHH379 pKa = 5.15GWKK382 pKa = 10.02PSDD385 pKa = 3.55ASAIPLIMSLYY396 pKa = 9.69NCLGLHH402 pKa = 5.43SQGKK406 pKa = 8.84SVYY409 pKa = 10.1KK410 pKa = 10.51LYY412 pKa = 10.4TVFQTLVKK420 pKa = 9.68TKK422 pKa = 9.52VTGEE426 pKa = 3.99YY427 pKa = 9.57STMAKK432 pKa = 10.65AMTEE436 pKa = 3.82AFRR439 pKa = 11.84WPFYY443 pKa = 8.21EE444 pKa = 3.93TSNEE448 pKa = 3.54IFASILGKK456 pKa = 9.84MKK458 pKa = 10.52EE459 pKa = 4.05EE460 pKa = 3.98QVWKK464 pKa = 10.71DD465 pKa = 3.45MEE467 pKa = 4.41SLQTNVYY474 pKa = 10.02LFHH477 pKa = 8.04DD478 pKa = 4.51EE479 pKa = 4.11FNKK482 pKa = 10.33RR483 pKa = 11.84MSTVNPLWASEE494 pKa = 4.4SYY496 pKa = 8.59KK497 pKa = 9.71TAYY500 pKa = 8.48TEE502 pKa = 3.95RR503 pKa = 11.84VPMLIMAEE511 pKa = 4.13YY512 pKa = 8.96LTMEE516 pKa = 4.58LEE518 pKa = 4.35EE519 pKa = 5.01KK520 pKa = 10.99LMILTGDD527 pKa = 3.66NSSDD531 pKa = 3.45EE532 pKa = 4.12YY533 pKa = 11.42QEE535 pKa = 5.79LIFNEE540 pKa = 4.23SLGSYY545 pKa = 9.84KK546 pKa = 10.4LSKK549 pKa = 10.65HH550 pKa = 5.26FFGLKK555 pKa = 10.35DD556 pKa = 3.41EE557 pKa = 4.83ASSVRR562 pKa = 11.84DD563 pKa = 3.14LSRR566 pKa = 11.84LVKK569 pKa = 10.1PLINKK574 pKa = 9.2VSEE577 pKa = 4.8IINKK581 pKa = 9.58SDD583 pKa = 3.13PTTSVILTTTEE594 pKa = 3.58QMFVKK599 pKa = 9.87DD600 pKa = 3.54TATFGPP606 pKa = 4.18
Molecular weight: 68.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
606 |
606 |
606 |
606.0 |
68.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.96 ± 0.0 | 0.825 ± 0.0 |
4.95 ± 0.0 | 5.941 ± 0.0 |
4.455 ± 0.0 | 5.611 ± 0.0 |
2.31 ± 0.0 | 5.446 ± 0.0 |
6.931 ± 0.0 | 9.736 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.95 ± 0.0 | 3.63 ± 0.0 |
4.29 ± 0.0 | 2.64 ± 0.0 |
2.31 ± 0.0 | 12.541 ± 0.0 |
7.096 ± 0.0 | 6.106 ± 0.0 |
2.97 ± 0.0 | 3.3 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
