
Thermus phage phiOH16
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; unclassified Inoviridae
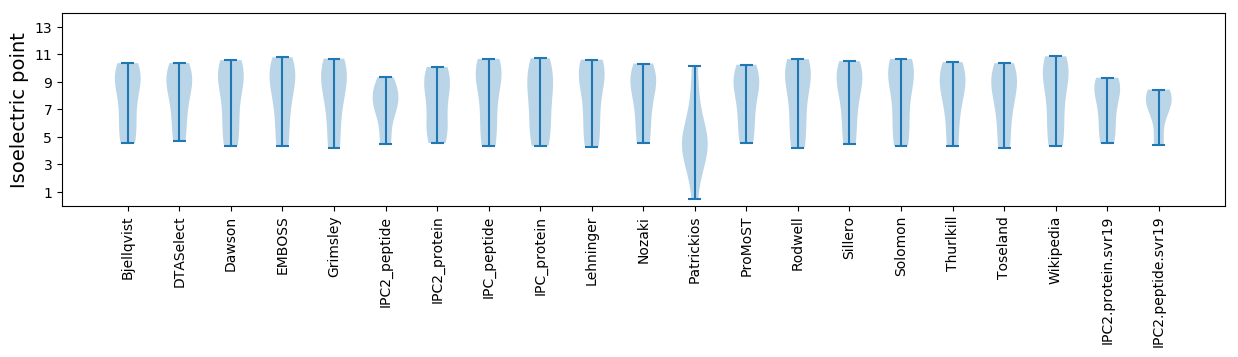
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
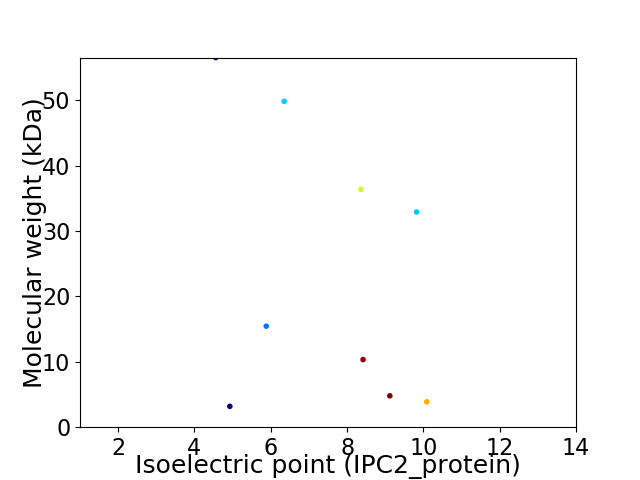
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S7J239|A0A1S7J239_9VIRU Minor coat protein OS=Thermus phage phiOH16 OX=1968340 GN=VI PE=4 SV=1
MM1 pKa = 7.43SRR3 pKa = 11.84LALPILLVLLSLGWGGPAFAQEE25 pKa = 4.27HH26 pKa = 5.76FKK28 pKa = 11.17GAIRR32 pKa = 11.84STVQGMTLDD41 pKa = 5.61EE42 pKa = 4.86IIQWSDD48 pKa = 2.68AQFRR52 pKa = 11.84RR53 pKa = 11.84QATIGRR59 pKa = 11.84YY60 pKa = 7.9VRR62 pKa = 11.84WGGAIVLGSALVYY75 pKa = 9.11TALDD79 pKa = 3.25YY80 pKa = 10.78FYY82 pKa = 11.29NVLKK86 pKa = 10.73AQTGTSLDD94 pKa = 2.93RR95 pKa = 11.84WYY97 pKa = 10.45FWSNPYY103 pKa = 10.18VEE105 pKa = 4.75VGRR108 pKa = 11.84CIDD111 pKa = 3.37RR112 pKa = 11.84GSYY115 pKa = 10.46RR116 pKa = 11.84DD117 pKa = 4.79FIAVAYY123 pKa = 10.47VNGRR127 pKa = 11.84QVNYY131 pKa = 10.16YY132 pKa = 8.52ATSHH136 pKa = 6.64FGKK139 pKa = 10.31CDD141 pKa = 3.49VPSLQSFTEE150 pKa = 4.08YY151 pKa = 9.95WYY153 pKa = 10.96GLHH156 pKa = 7.07LVRR159 pKa = 11.84NYY161 pKa = 11.07SEE163 pKa = 4.26LQGVPTQYY171 pKa = 10.77VSPPPGTCPSGWTCIAVAPQLDD193 pKa = 4.49RR194 pKa = 11.84PPLPDD199 pKa = 3.39WLQSHH204 pKa = 7.6PDD206 pKa = 3.16AADD209 pKa = 3.53GVKK212 pKa = 10.1QAVTTYY218 pKa = 11.07LDD220 pKa = 3.8RR221 pKa = 11.84NPIGSPYY228 pKa = 10.58APYY231 pKa = 10.19PGVQLEE237 pKa = 4.97PIPNPNQWTDD247 pKa = 3.29NPFTRR252 pKa = 11.84PDD254 pKa = 3.46IDD256 pKa = 3.52TDD258 pKa = 3.77GDD260 pKa = 3.69GWPDD264 pKa = 3.31PVEE267 pKa = 3.51WRR269 pKa = 11.84EE270 pKa = 3.72ANRR273 pKa = 11.84RR274 pKa = 11.84GVPWPDD280 pKa = 5.02LINNPQAYY288 pKa = 7.88PDD290 pKa = 4.32PNGDD294 pKa = 3.58PDD296 pKa = 4.77GDD298 pKa = 3.67GFTNLEE304 pKa = 4.12EE305 pKa = 4.42VQQGTDD311 pKa = 3.17PYY313 pKa = 10.74DD314 pKa = 3.9PTSHH318 pKa = 7.06PVRR321 pKa = 11.84RR322 pKa = 11.84SPTSPWVDD330 pKa = 3.05TDD332 pKa = 3.34GDD334 pKa = 4.29GYY336 pKa = 11.18SDD338 pKa = 3.59EE339 pKa = 4.36EE340 pKa = 4.49EE341 pKa = 3.98IRR343 pKa = 11.84KK344 pKa = 7.8GTNPNDD350 pKa = 3.54PASYY354 pKa = 10.12PDD356 pKa = 4.25TPPEE360 pKa = 3.93QQPEE364 pKa = 4.26EE365 pKa = 4.51NPDD368 pKa = 3.22EE369 pKa = 4.5PQWPGGPPSGRR380 pKa = 11.84IDD382 pKa = 3.4PVQLPEE388 pKa = 3.98VEE390 pKa = 4.04QVEE393 pKa = 4.5RR394 pKa = 11.84EE395 pKa = 4.33KK396 pKa = 11.31LPEE399 pKa = 3.83WDD401 pKa = 3.96KK402 pKa = 11.71LNPLAEE408 pKa = 4.1AWRR411 pKa = 11.84QQVVDD416 pKa = 3.9RR417 pKa = 11.84VSQKK421 pKa = 10.05FAEE424 pKa = 4.35VQNILKK430 pKa = 10.46DD431 pKa = 3.55RR432 pKa = 11.84FPFGIIAAIRR442 pKa = 11.84QRR444 pKa = 11.84VSFADD449 pKa = 3.68AQCAIQLSLPPLGTLAVDD467 pKa = 3.84ICSTPVWQLATSFRR481 pKa = 11.84PVLAGLVWVAFGFAVIRR498 pKa = 11.84RR499 pKa = 11.84ALDD502 pKa = 3.3VQKK505 pKa = 11.3
MM1 pKa = 7.43SRR3 pKa = 11.84LALPILLVLLSLGWGGPAFAQEE25 pKa = 4.27HH26 pKa = 5.76FKK28 pKa = 11.17GAIRR32 pKa = 11.84STVQGMTLDD41 pKa = 5.61EE42 pKa = 4.86IIQWSDD48 pKa = 2.68AQFRR52 pKa = 11.84RR53 pKa = 11.84QATIGRR59 pKa = 11.84YY60 pKa = 7.9VRR62 pKa = 11.84WGGAIVLGSALVYY75 pKa = 9.11TALDD79 pKa = 3.25YY80 pKa = 10.78FYY82 pKa = 11.29NVLKK86 pKa = 10.73AQTGTSLDD94 pKa = 2.93RR95 pKa = 11.84WYY97 pKa = 10.45FWSNPYY103 pKa = 10.18VEE105 pKa = 4.75VGRR108 pKa = 11.84CIDD111 pKa = 3.37RR112 pKa = 11.84GSYY115 pKa = 10.46RR116 pKa = 11.84DD117 pKa = 4.79FIAVAYY123 pKa = 10.47VNGRR127 pKa = 11.84QVNYY131 pKa = 10.16YY132 pKa = 8.52ATSHH136 pKa = 6.64FGKK139 pKa = 10.31CDD141 pKa = 3.49VPSLQSFTEE150 pKa = 4.08YY151 pKa = 9.95WYY153 pKa = 10.96GLHH156 pKa = 7.07LVRR159 pKa = 11.84NYY161 pKa = 11.07SEE163 pKa = 4.26LQGVPTQYY171 pKa = 10.77VSPPPGTCPSGWTCIAVAPQLDD193 pKa = 4.49RR194 pKa = 11.84PPLPDD199 pKa = 3.39WLQSHH204 pKa = 7.6PDD206 pKa = 3.16AADD209 pKa = 3.53GVKK212 pKa = 10.1QAVTTYY218 pKa = 11.07LDD220 pKa = 3.8RR221 pKa = 11.84NPIGSPYY228 pKa = 10.58APYY231 pKa = 10.19PGVQLEE237 pKa = 4.97PIPNPNQWTDD247 pKa = 3.29NPFTRR252 pKa = 11.84PDD254 pKa = 3.46IDD256 pKa = 3.52TDD258 pKa = 3.77GDD260 pKa = 3.69GWPDD264 pKa = 3.31PVEE267 pKa = 3.51WRR269 pKa = 11.84EE270 pKa = 3.72ANRR273 pKa = 11.84RR274 pKa = 11.84GVPWPDD280 pKa = 5.02LINNPQAYY288 pKa = 7.88PDD290 pKa = 4.32PNGDD294 pKa = 3.58PDD296 pKa = 4.77GDD298 pKa = 3.67GFTNLEE304 pKa = 4.12EE305 pKa = 4.42VQQGTDD311 pKa = 3.17PYY313 pKa = 10.74DD314 pKa = 3.9PTSHH318 pKa = 7.06PVRR321 pKa = 11.84RR322 pKa = 11.84SPTSPWVDD330 pKa = 3.05TDD332 pKa = 3.34GDD334 pKa = 4.29GYY336 pKa = 11.18SDD338 pKa = 3.59EE339 pKa = 4.36EE340 pKa = 4.49EE341 pKa = 3.98IRR343 pKa = 11.84KK344 pKa = 7.8GTNPNDD350 pKa = 3.54PASYY354 pKa = 10.12PDD356 pKa = 4.25TPPEE360 pKa = 3.93QQPEE364 pKa = 4.26EE365 pKa = 4.51NPDD368 pKa = 3.22EE369 pKa = 4.5PQWPGGPPSGRR380 pKa = 11.84IDD382 pKa = 3.4PVQLPEE388 pKa = 3.98VEE390 pKa = 4.04QVEE393 pKa = 4.5RR394 pKa = 11.84EE395 pKa = 4.33KK396 pKa = 11.31LPEE399 pKa = 3.83WDD401 pKa = 3.96KK402 pKa = 11.71LNPLAEE408 pKa = 4.1AWRR411 pKa = 11.84QQVVDD416 pKa = 3.9RR417 pKa = 11.84VSQKK421 pKa = 10.05FAEE424 pKa = 4.35VQNILKK430 pKa = 10.46DD431 pKa = 3.55RR432 pKa = 11.84FPFGIIAAIRR442 pKa = 11.84QRR444 pKa = 11.84VSFADD449 pKa = 3.68AQCAIQLSLPPLGTLAVDD467 pKa = 3.84ICSTPVWQLATSFRR481 pKa = 11.84PVLAGLVWVAFGFAVIRR498 pKa = 11.84RR499 pKa = 11.84ALDD502 pKa = 3.3VQKK505 pKa = 11.3
Molecular weight: 56.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S7J233|A0A1S7J233_9VIRU Replication endonuclease OS=Thermus phage phiOH16 OX=1968340 GN=II PE=4 SV=1
MM1 pKa = 7.92DD2 pKa = 4.46FVGVDD7 pKa = 3.69VSAKK11 pKa = 10.25RR12 pKa = 11.84LDD14 pKa = 3.47VALGGKK20 pKa = 9.25LLSFPNPEE28 pKa = 4.87GIPDD32 pKa = 5.34LIAALPPGAVVGLEE46 pKa = 3.73ATGVYY51 pKa = 9.63GRR53 pKa = 11.84PLAFALHH60 pKa = 6.37RR61 pKa = 11.84AGFHH65 pKa = 6.18VYY67 pKa = 9.79VLNPFAVKK75 pKa = 10.18SYY77 pKa = 10.94ARR79 pKa = 11.84SLLLRR84 pKa = 11.84SKK86 pKa = 9.48TDD88 pKa = 3.15RR89 pKa = 11.84VDD91 pKa = 2.92ARR93 pKa = 11.84LIARR97 pKa = 11.84YY98 pKa = 8.75LADD101 pKa = 5.26RR102 pKa = 11.84YY103 pKa = 10.2PDD105 pKa = 3.52LPLYY109 pKa = 10.24EE110 pKa = 4.63PAPEE114 pKa = 4.28VIYY117 pKa = 10.57QAGLLVRR124 pKa = 11.84FARR127 pKa = 11.84GLVVQRR133 pKa = 11.84VAVLNRR139 pKa = 11.84LHH141 pKa = 6.74AWQYY145 pKa = 10.6AWPQGLEE152 pKa = 4.35VVAHH156 pKa = 5.65VPRR159 pKa = 11.84TLEE162 pKa = 3.76ALRR165 pKa = 11.84GNLEE169 pKa = 3.95ALAVDD174 pKa = 5.05LLRR177 pKa = 11.84SDD179 pKa = 3.9PEE181 pKa = 3.29AWRR184 pKa = 11.84QFRR187 pKa = 11.84ALQTLPGIGPSSALTILAYY206 pKa = 10.7SGDD209 pKa = 3.32LRR211 pKa = 11.84RR212 pKa = 11.84FKK214 pKa = 10.7SARR217 pKa = 11.84AYY219 pKa = 10.54AAFTGLAPVVHH230 pKa = 6.12QSGALPEE237 pKa = 3.98VGRR240 pKa = 11.84ISRR243 pKa = 11.84AGPAPLRR250 pKa = 11.84ATFWLASLHH259 pKa = 5.32AVKK262 pKa = 10.69VEE264 pKa = 4.65PYY266 pKa = 9.19ASLYY270 pKa = 10.91ARR272 pKa = 11.84LLQAGKK278 pKa = 8.58PPQVARR284 pKa = 11.84VAVAHH289 pKa = 6.35RR290 pKa = 11.84LARR293 pKa = 11.84AAWAVVVRR301 pKa = 11.84SVV303 pKa = 3.24
MM1 pKa = 7.92DD2 pKa = 4.46FVGVDD7 pKa = 3.69VSAKK11 pKa = 10.25RR12 pKa = 11.84LDD14 pKa = 3.47VALGGKK20 pKa = 9.25LLSFPNPEE28 pKa = 4.87GIPDD32 pKa = 5.34LIAALPPGAVVGLEE46 pKa = 3.73ATGVYY51 pKa = 9.63GRR53 pKa = 11.84PLAFALHH60 pKa = 6.37RR61 pKa = 11.84AGFHH65 pKa = 6.18VYY67 pKa = 9.79VLNPFAVKK75 pKa = 10.18SYY77 pKa = 10.94ARR79 pKa = 11.84SLLLRR84 pKa = 11.84SKK86 pKa = 9.48TDD88 pKa = 3.15RR89 pKa = 11.84VDD91 pKa = 2.92ARR93 pKa = 11.84LIARR97 pKa = 11.84YY98 pKa = 8.75LADD101 pKa = 5.26RR102 pKa = 11.84YY103 pKa = 10.2PDD105 pKa = 3.52LPLYY109 pKa = 10.24EE110 pKa = 4.63PAPEE114 pKa = 4.28VIYY117 pKa = 10.57QAGLLVRR124 pKa = 11.84FARR127 pKa = 11.84GLVVQRR133 pKa = 11.84VAVLNRR139 pKa = 11.84LHH141 pKa = 6.74AWQYY145 pKa = 10.6AWPQGLEE152 pKa = 4.35VVAHH156 pKa = 5.65VPRR159 pKa = 11.84TLEE162 pKa = 3.76ALRR165 pKa = 11.84GNLEE169 pKa = 3.95ALAVDD174 pKa = 5.05LLRR177 pKa = 11.84SDD179 pKa = 3.9PEE181 pKa = 3.29AWRR184 pKa = 11.84QFRR187 pKa = 11.84ALQTLPGIGPSSALTILAYY206 pKa = 10.7SGDD209 pKa = 3.32LRR211 pKa = 11.84RR212 pKa = 11.84FKK214 pKa = 10.7SARR217 pKa = 11.84AYY219 pKa = 10.54AAFTGLAPVVHH230 pKa = 6.12QSGALPEE237 pKa = 3.98VGRR240 pKa = 11.84ISRR243 pKa = 11.84AGPAPLRR250 pKa = 11.84ATFWLASLHH259 pKa = 5.32AVKK262 pKa = 10.69VEE264 pKa = 4.65PYY266 pKa = 9.19ASLYY270 pKa = 10.91ARR272 pKa = 11.84LLQAGKK278 pKa = 8.58PPQVARR284 pKa = 11.84VAVAHH289 pKa = 6.35RR290 pKa = 11.84LARR293 pKa = 11.84AAWAVVVRR301 pKa = 11.84SVV303 pKa = 3.24
Molecular weight: 32.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1921 |
30 |
505 |
213.4 |
23.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.984 ± 1.497 | 0.469 ± 0.262 |
5.362 ± 0.817 | 4.945 ± 0.742 |
3.852 ± 0.288 | 7.86 ± 0.493 |
1.51 ± 0.251 | 3.644 ± 0.804 |
2.811 ± 0.455 | 10.828 ± 1.499 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.406 ± 0.411 | 2.134 ± 0.507 |
7.236 ± 1.129 | 3.54 ± 0.853 |
7.496 ± 0.791 | 5.414 ± 0.364 |
3.8 ± 0.526 | 9.682 ± 0.856 |
3.227 ± 0.446 | 3.8 ± 0.332 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
