
Human coronavirus 229E (HCoV-229E)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Duvinacovirus
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|P19739|NS4A_CVH22 Non-structural protein 4a OS=Human coronavirus 229E OX=11137 GN=4a PE=3 SV=1
MM1 pKa = 7.16FVLLVAYY8 pKa = 9.84ALLHH12 pKa = 6.24IAGCQTTNGLNTSYY26 pKa = 10.55SVCNGCVGYY35 pKa = 10.09SEE37 pKa = 5.49NVFAVEE43 pKa = 4.01SGGYY47 pKa = 9.24IPSDD51 pKa = 3.14FAFNNWFLLTNTSSVVDD68 pKa = 3.68GVVRR72 pKa = 11.84SFQPLLLNCLWSVSGLRR89 pKa = 11.84FTTGFVYY96 pKa = 10.41FNGTGRR102 pKa = 11.84GDD104 pKa = 3.68CKK106 pKa = 10.91GFSSDD111 pKa = 3.57VLSDD115 pKa = 3.58VIRR118 pKa = 11.84YY119 pKa = 8.08NLNFEE124 pKa = 4.23EE125 pKa = 4.14NLRR128 pKa = 11.84RR129 pKa = 11.84GTILFKK135 pKa = 10.17TSYY138 pKa = 9.79GVVVFYY144 pKa = 8.57CTNNTLVSGDD154 pKa = 3.18AHH156 pKa = 7.49IPFGTVLGNFYY167 pKa = 10.84CFVNTTIGNEE177 pKa = 4.05TTSAFVGALPKK188 pKa = 9.64TVRR191 pKa = 11.84EE192 pKa = 4.1FVISRR197 pKa = 11.84TGHH200 pKa = 6.13FYY202 pKa = 10.78INGYY206 pKa = 10.46RR207 pKa = 11.84YY208 pKa = 7.33FTLGNVEE215 pKa = 3.93AVNFNVTTAEE225 pKa = 4.07TTDD228 pKa = 3.8FCTVALASYY237 pKa = 11.29ADD239 pKa = 3.47VLVNVSQTSIANIIYY254 pKa = 9.55CNSVINRR261 pKa = 11.84LRR263 pKa = 11.84CDD265 pKa = 3.35QLSFDD270 pKa = 4.48VPDD273 pKa = 4.15GFYY276 pKa = 9.65STSPIQSVEE285 pKa = 4.08LPVSIVSLPVYY296 pKa = 9.8HH297 pKa = 6.85KK298 pKa = 9.52HH299 pKa = 4.94TFIVLYY305 pKa = 10.81VDD307 pKa = 5.79FKK309 pKa = 10.03PQSGGGKK316 pKa = 9.91CFNCYY321 pKa = 8.96PAGVNITLANFNEE334 pKa = 4.85TKK336 pKa = 10.79GPLCVDD342 pKa = 4.0TSHH345 pKa = 6.91FTTKK349 pKa = 10.36YY350 pKa = 7.47VAVYY354 pKa = 10.46ANVGRR359 pKa = 11.84WSASINTGNCPFSFGKK375 pKa = 10.56VNNFVKK381 pKa = 10.67FGSVCFSLKK390 pKa = 10.45DD391 pKa = 3.56IPGGCAMPIVANWAYY406 pKa = 10.72SKK408 pKa = 11.02YY409 pKa = 8.53YY410 pKa = 10.08TIGSLYY416 pKa = 10.64VSWSDD421 pKa = 3.16GDD423 pKa = 4.5GITGVPQPVEE433 pKa = 4.29GVSSFMNVTLDD444 pKa = 2.87KK445 pKa = 9.64CTKK448 pKa = 9.1YY449 pKa = 10.83NIYY452 pKa = 10.19DD453 pKa = 3.42VSGVGVIRR461 pKa = 11.84VSNDD465 pKa = 2.51TFLNGITYY473 pKa = 9.16TSTSGNLLGFKK484 pKa = 10.26DD485 pKa = 3.78VTKK488 pKa = 9.86GTIYY492 pKa = 10.61SITPCNPPDD501 pKa = 3.49QLVVYY506 pKa = 8.2QQAVVGAMLSEE517 pKa = 4.28NFTSYY522 pKa = 11.02GFSNVVEE529 pKa = 4.31LPKK532 pKa = 10.59FFYY535 pKa = 10.75ASNGTYY541 pKa = 10.3NCTDD545 pKa = 2.86AVLTYY550 pKa = 11.05SSFGVCADD558 pKa = 3.43GSIIAVQPRR567 pKa = 11.84NVSYY571 pKa = 11.0DD572 pKa = 3.48SVSAIVTANLSIPSNWTTSVQVEE595 pKa = 4.12YY596 pKa = 10.83LQITSTPIVVDD607 pKa = 3.85CSTYY611 pKa = 9.47VCNGNVRR618 pKa = 11.84CVEE621 pKa = 4.02LLKK624 pKa = 10.78QYY626 pKa = 10.53TSACKK631 pKa = 9.83TIEE634 pKa = 4.11DD635 pKa = 4.03ALRR638 pKa = 11.84NSARR642 pKa = 11.84LEE644 pKa = 4.09SADD647 pKa = 3.53VSEE650 pKa = 5.05MLTFDD655 pKa = 3.73KK656 pKa = 11.09KK657 pKa = 11.1AFTLANVSSFGDD669 pKa = 3.69YY670 pKa = 10.6NLSSVIPSLPTSGSRR685 pKa = 11.84VAGRR689 pKa = 11.84SAIEE693 pKa = 5.09DD694 pKa = 3.32ILFSKK699 pKa = 10.7LVTSGLGTVDD709 pKa = 4.5ADD711 pKa = 3.86YY712 pKa = 11.22KK713 pKa = 11.03KK714 pKa = 9.54CTKK717 pKa = 10.13GLSIADD723 pKa = 4.44LACAQYY729 pKa = 11.34YY730 pKa = 9.2NGIMVLPGVADD741 pKa = 4.01AEE743 pKa = 4.12RR744 pKa = 11.84MAMYY748 pKa = 9.17TGSLIGGIALGGLTSAVSIPFSLAIQARR776 pKa = 11.84LNYY779 pKa = 9.7VALQTDD785 pKa = 4.22VLQEE789 pKa = 3.94NQKK792 pKa = 9.9ILAASFNKK800 pKa = 10.34AMTNIVDD807 pKa = 3.87AFTGVNDD814 pKa = 6.2AITQTSQALQTVATALNKK832 pKa = 10.09IQDD835 pKa = 3.83VVNQQGNSLNHH846 pKa = 6.06LTSQLRR852 pKa = 11.84QNFQAISSSIQAIYY866 pKa = 10.75DD867 pKa = 3.62RR868 pKa = 11.84LDD870 pKa = 3.71TIQADD875 pKa = 3.81QQVDD879 pKa = 3.53RR880 pKa = 11.84LITGRR885 pKa = 11.84LAALNVFVSHH895 pKa = 7.02TLTKK899 pKa = 8.3YY900 pKa = 8.64TEE902 pKa = 4.22VRR904 pKa = 11.84ASRR907 pKa = 11.84QLAQQKK913 pKa = 10.1VNEE916 pKa = 4.44CVKK919 pKa = 10.49SQSKK923 pKa = 10.49RR924 pKa = 11.84YY925 pKa = 8.36GFCGNGTHH933 pKa = 6.96IFSIVNAAPEE943 pKa = 4.27GLVFLHH949 pKa = 6.06TVLLPTQYY957 pKa = 11.19KK958 pKa = 10.38DD959 pKa = 3.27VEE961 pKa = 4.35AWSGLCVDD969 pKa = 4.6GTNGYY974 pKa = 7.64VLRR977 pKa = 11.84QPNLALYY984 pKa = 10.59KK985 pKa = 9.95EE986 pKa = 4.32GNYY989 pKa = 10.11YY990 pKa = 10.72RR991 pKa = 11.84ITSRR995 pKa = 11.84IMFEE999 pKa = 3.71PRR1001 pKa = 11.84IPTMADD1007 pKa = 3.54FVQIEE1012 pKa = 4.38NCNVTFVNISRR1023 pKa = 11.84SEE1025 pKa = 3.96LQTIVPEE1032 pKa = 4.33YY1033 pKa = 10.5IDD1035 pKa = 3.57VNKK1038 pKa = 9.33TLQEE1042 pKa = 3.95LSYY1045 pKa = 10.83KK1046 pKa = 10.05LPNYY1050 pKa = 7.31TVPDD1054 pKa = 3.84LVVEE1058 pKa = 4.42QYY1060 pKa = 11.18NQTILNLTSEE1070 pKa = 4.16ISTLEE1075 pKa = 4.02NKK1077 pKa = 9.98SAEE1080 pKa = 4.16LNYY1083 pKa = 9.73TVQKK1087 pKa = 10.55LQTLIDD1093 pKa = 4.68NINSTLVDD1101 pKa = 4.5LKK1103 pKa = 10.17WLNRR1107 pKa = 11.84VEE1109 pKa = 5.36TYY1111 pKa = 10.41IKK1113 pKa = 9.23WPWWVWLCISVVLIFVVSMLLLCCCSTGCCGFFSCFASSIRR1154 pKa = 11.84GCCEE1158 pKa = 3.71STKK1161 pKa = 10.6LPYY1164 pKa = 10.38YY1165 pKa = 10.29DD1166 pKa = 3.56VEE1168 pKa = 5.49KK1169 pKa = 10.86IHH1171 pKa = 6.47IQQ1173 pKa = 3.23
MM1 pKa = 7.16FVLLVAYY8 pKa = 9.84ALLHH12 pKa = 6.24IAGCQTTNGLNTSYY26 pKa = 10.55SVCNGCVGYY35 pKa = 10.09SEE37 pKa = 5.49NVFAVEE43 pKa = 4.01SGGYY47 pKa = 9.24IPSDD51 pKa = 3.14FAFNNWFLLTNTSSVVDD68 pKa = 3.68GVVRR72 pKa = 11.84SFQPLLLNCLWSVSGLRR89 pKa = 11.84FTTGFVYY96 pKa = 10.41FNGTGRR102 pKa = 11.84GDD104 pKa = 3.68CKK106 pKa = 10.91GFSSDD111 pKa = 3.57VLSDD115 pKa = 3.58VIRR118 pKa = 11.84YY119 pKa = 8.08NLNFEE124 pKa = 4.23EE125 pKa = 4.14NLRR128 pKa = 11.84RR129 pKa = 11.84GTILFKK135 pKa = 10.17TSYY138 pKa = 9.79GVVVFYY144 pKa = 8.57CTNNTLVSGDD154 pKa = 3.18AHH156 pKa = 7.49IPFGTVLGNFYY167 pKa = 10.84CFVNTTIGNEE177 pKa = 4.05TTSAFVGALPKK188 pKa = 9.64TVRR191 pKa = 11.84EE192 pKa = 4.1FVISRR197 pKa = 11.84TGHH200 pKa = 6.13FYY202 pKa = 10.78INGYY206 pKa = 10.46RR207 pKa = 11.84YY208 pKa = 7.33FTLGNVEE215 pKa = 3.93AVNFNVTTAEE225 pKa = 4.07TTDD228 pKa = 3.8FCTVALASYY237 pKa = 11.29ADD239 pKa = 3.47VLVNVSQTSIANIIYY254 pKa = 9.55CNSVINRR261 pKa = 11.84LRR263 pKa = 11.84CDD265 pKa = 3.35QLSFDD270 pKa = 4.48VPDD273 pKa = 4.15GFYY276 pKa = 9.65STSPIQSVEE285 pKa = 4.08LPVSIVSLPVYY296 pKa = 9.8HH297 pKa = 6.85KK298 pKa = 9.52HH299 pKa = 4.94TFIVLYY305 pKa = 10.81VDD307 pKa = 5.79FKK309 pKa = 10.03PQSGGGKK316 pKa = 9.91CFNCYY321 pKa = 8.96PAGVNITLANFNEE334 pKa = 4.85TKK336 pKa = 10.79GPLCVDD342 pKa = 4.0TSHH345 pKa = 6.91FTTKK349 pKa = 10.36YY350 pKa = 7.47VAVYY354 pKa = 10.46ANVGRR359 pKa = 11.84WSASINTGNCPFSFGKK375 pKa = 10.56VNNFVKK381 pKa = 10.67FGSVCFSLKK390 pKa = 10.45DD391 pKa = 3.56IPGGCAMPIVANWAYY406 pKa = 10.72SKK408 pKa = 11.02YY409 pKa = 8.53YY410 pKa = 10.08TIGSLYY416 pKa = 10.64VSWSDD421 pKa = 3.16GDD423 pKa = 4.5GITGVPQPVEE433 pKa = 4.29GVSSFMNVTLDD444 pKa = 2.87KK445 pKa = 9.64CTKK448 pKa = 9.1YY449 pKa = 10.83NIYY452 pKa = 10.19DD453 pKa = 3.42VSGVGVIRR461 pKa = 11.84VSNDD465 pKa = 2.51TFLNGITYY473 pKa = 9.16TSTSGNLLGFKK484 pKa = 10.26DD485 pKa = 3.78VTKK488 pKa = 9.86GTIYY492 pKa = 10.61SITPCNPPDD501 pKa = 3.49QLVVYY506 pKa = 8.2QQAVVGAMLSEE517 pKa = 4.28NFTSYY522 pKa = 11.02GFSNVVEE529 pKa = 4.31LPKK532 pKa = 10.59FFYY535 pKa = 10.75ASNGTYY541 pKa = 10.3NCTDD545 pKa = 2.86AVLTYY550 pKa = 11.05SSFGVCADD558 pKa = 3.43GSIIAVQPRR567 pKa = 11.84NVSYY571 pKa = 11.0DD572 pKa = 3.48SVSAIVTANLSIPSNWTTSVQVEE595 pKa = 4.12YY596 pKa = 10.83LQITSTPIVVDD607 pKa = 3.85CSTYY611 pKa = 9.47VCNGNVRR618 pKa = 11.84CVEE621 pKa = 4.02LLKK624 pKa = 10.78QYY626 pKa = 10.53TSACKK631 pKa = 9.83TIEE634 pKa = 4.11DD635 pKa = 4.03ALRR638 pKa = 11.84NSARR642 pKa = 11.84LEE644 pKa = 4.09SADD647 pKa = 3.53VSEE650 pKa = 5.05MLTFDD655 pKa = 3.73KK656 pKa = 11.09KK657 pKa = 11.1AFTLANVSSFGDD669 pKa = 3.69YY670 pKa = 10.6NLSSVIPSLPTSGSRR685 pKa = 11.84VAGRR689 pKa = 11.84SAIEE693 pKa = 5.09DD694 pKa = 3.32ILFSKK699 pKa = 10.7LVTSGLGTVDD709 pKa = 4.5ADD711 pKa = 3.86YY712 pKa = 11.22KK713 pKa = 11.03KK714 pKa = 9.54CTKK717 pKa = 10.13GLSIADD723 pKa = 4.44LACAQYY729 pKa = 11.34YY730 pKa = 9.2NGIMVLPGVADD741 pKa = 4.01AEE743 pKa = 4.12RR744 pKa = 11.84MAMYY748 pKa = 9.17TGSLIGGIALGGLTSAVSIPFSLAIQARR776 pKa = 11.84LNYY779 pKa = 9.7VALQTDD785 pKa = 4.22VLQEE789 pKa = 3.94NQKK792 pKa = 9.9ILAASFNKK800 pKa = 10.34AMTNIVDD807 pKa = 3.87AFTGVNDD814 pKa = 6.2AITQTSQALQTVATALNKK832 pKa = 10.09IQDD835 pKa = 3.83VVNQQGNSLNHH846 pKa = 6.06LTSQLRR852 pKa = 11.84QNFQAISSSIQAIYY866 pKa = 10.75DD867 pKa = 3.62RR868 pKa = 11.84LDD870 pKa = 3.71TIQADD875 pKa = 3.81QQVDD879 pKa = 3.53RR880 pKa = 11.84LITGRR885 pKa = 11.84LAALNVFVSHH895 pKa = 7.02TLTKK899 pKa = 8.3YY900 pKa = 8.64TEE902 pKa = 4.22VRR904 pKa = 11.84ASRR907 pKa = 11.84QLAQQKK913 pKa = 10.1VNEE916 pKa = 4.44CVKK919 pKa = 10.49SQSKK923 pKa = 10.49RR924 pKa = 11.84YY925 pKa = 8.36GFCGNGTHH933 pKa = 6.96IFSIVNAAPEE943 pKa = 4.27GLVFLHH949 pKa = 6.06TVLLPTQYY957 pKa = 11.19KK958 pKa = 10.38DD959 pKa = 3.27VEE961 pKa = 4.35AWSGLCVDD969 pKa = 4.6GTNGYY974 pKa = 7.64VLRR977 pKa = 11.84QPNLALYY984 pKa = 10.59KK985 pKa = 9.95EE986 pKa = 4.32GNYY989 pKa = 10.11YY990 pKa = 10.72RR991 pKa = 11.84ITSRR995 pKa = 11.84IMFEE999 pKa = 3.71PRR1001 pKa = 11.84IPTMADD1007 pKa = 3.54FVQIEE1012 pKa = 4.38NCNVTFVNISRR1023 pKa = 11.84SEE1025 pKa = 3.96LQTIVPEE1032 pKa = 4.33YY1033 pKa = 10.5IDD1035 pKa = 3.57VNKK1038 pKa = 9.33TLQEE1042 pKa = 3.95LSYY1045 pKa = 10.83KK1046 pKa = 10.05LPNYY1050 pKa = 7.31TVPDD1054 pKa = 3.84LVVEE1058 pKa = 4.42QYY1060 pKa = 11.18NQTILNLTSEE1070 pKa = 4.16ISTLEE1075 pKa = 4.02NKK1077 pKa = 9.98SAEE1080 pKa = 4.16LNYY1083 pKa = 9.73TVQKK1087 pKa = 10.55LQTLIDD1093 pKa = 4.68NINSTLVDD1101 pKa = 4.5LKK1103 pKa = 10.17WLNRR1107 pKa = 11.84VEE1109 pKa = 5.36TYY1111 pKa = 10.41IKK1113 pKa = 9.23WPWWVWLCISVVLIFVVSMLLLCCCSTGCCGFFSCFASSIRR1154 pKa = 11.84GCCEE1158 pKa = 3.71STKK1161 pKa = 10.6LPYY1164 pKa = 10.38YY1165 pKa = 10.29DD1166 pKa = 3.56VEE1168 pKa = 5.49KK1169 pKa = 10.86IHH1171 pKa = 6.47IQQ1173 pKa = 3.23
Molecular weight: 128.64 kDa
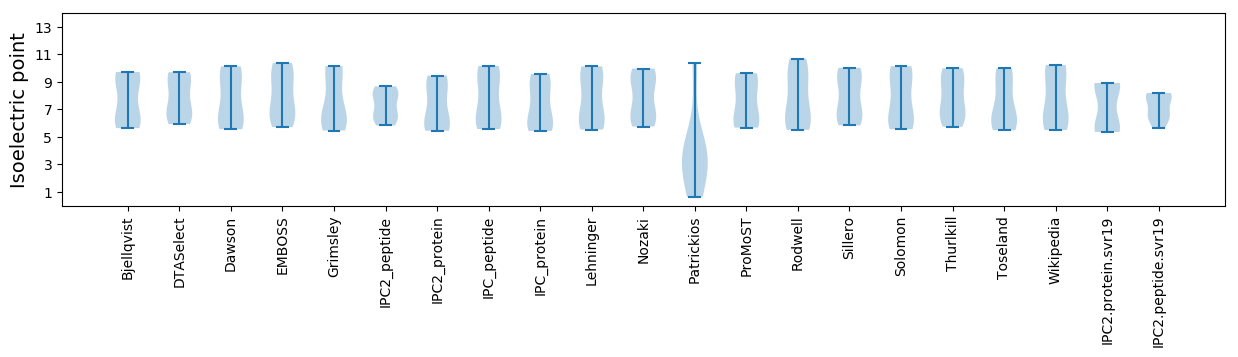
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P15422|VME1_CVH22 Membrane protein OS=Human coronavirus 229E OX=11137 GN=M PE=3 SV=3
MM1 pKa = 7.31ATVKK5 pKa = 9.76WADD8 pKa = 2.98ASEE11 pKa = 4.15PQRR14 pKa = 11.84GRR16 pKa = 11.84QGRR19 pKa = 11.84IPYY22 pKa = 9.97SLYY25 pKa = 10.99SPLLVDD31 pKa = 5.73SEE33 pKa = 4.75QPWKK37 pKa = 10.7VIPRR41 pKa = 11.84NLVPINKK48 pKa = 8.93KK49 pKa = 10.4DD50 pKa = 3.45KK51 pKa = 11.0NKK53 pKa = 10.73LIGYY57 pKa = 7.6WNVQKK62 pKa = 10.81RR63 pKa = 11.84FRR65 pKa = 11.84TRR67 pKa = 11.84KK68 pKa = 8.8GKK70 pKa = 10.37RR71 pKa = 11.84VDD73 pKa = 4.23LSPKK77 pKa = 9.28LHH79 pKa = 6.82FYY81 pKa = 11.19YY82 pKa = 10.7LGTGPHH88 pKa = 6.76KK89 pKa = 10.21DD90 pKa = 2.84AKK92 pKa = 10.2FRR94 pKa = 11.84EE95 pKa = 4.43RR96 pKa = 11.84VEE98 pKa = 4.13GVVWVAVDD106 pKa = 3.79GAKK109 pKa = 9.47TEE111 pKa = 3.99PTGYY115 pKa = 10.26GVRR118 pKa = 11.84RR119 pKa = 11.84KK120 pKa = 10.01NSEE123 pKa = 3.64PEE125 pKa = 3.74IPHH128 pKa = 6.81FNQKK132 pKa = 9.87LPNGVTVVEE141 pKa = 4.8EE142 pKa = 4.2PDD144 pKa = 3.07SRR146 pKa = 11.84APSRR150 pKa = 11.84SQSRR154 pKa = 11.84SQSRR158 pKa = 11.84GRR160 pKa = 11.84GEE162 pKa = 4.47SKK164 pKa = 9.89PQSRR168 pKa = 11.84NPSSDD173 pKa = 3.46RR174 pKa = 11.84NHH176 pKa = 6.45NSQDD180 pKa = 3.98DD181 pKa = 3.55IMKK184 pKa = 10.59AVAAALKK191 pKa = 10.67SLGFDD196 pKa = 3.67KK197 pKa = 10.89PQEE200 pKa = 4.05KK201 pKa = 10.13DD202 pKa = 3.04KK203 pKa = 11.44KK204 pKa = 9.11SAKK207 pKa = 9.64TGTPKK212 pKa = 10.47PSRR215 pKa = 11.84NQSPASSQTSAKK227 pKa = 10.2SLARR231 pKa = 11.84SQSSEE236 pKa = 3.88TKK238 pKa = 7.93EE239 pKa = 4.32QKK241 pKa = 10.46HH242 pKa = 5.47EE243 pKa = 4.05MQKK246 pKa = 10.2PRR248 pKa = 11.84WKK250 pKa = 10.22RR251 pKa = 11.84QPNDD255 pKa = 3.49DD256 pKa = 3.43VTSNVTQCFGPRR268 pKa = 11.84DD269 pKa = 3.6LDD271 pKa = 3.95HH272 pKa = 7.0NFGSAGVVANGVKK285 pKa = 10.53AKK287 pKa = 10.29GYY289 pKa = 8.25PQFAEE294 pKa = 4.56LVPSTAAMLFDD305 pKa = 3.29SHH307 pKa = 7.6IVSKK311 pKa = 10.89EE312 pKa = 3.77SGNTVVLTFTTRR324 pKa = 11.84VTVPKK329 pKa = 10.31DD330 pKa = 3.33HH331 pKa = 6.76PHH333 pKa = 6.16LGKK336 pKa = 10.32FLEE339 pKa = 4.34EE340 pKa = 3.99LNAFTRR346 pKa = 11.84EE347 pKa = 4.19MQQHH351 pKa = 6.7PLLNPSALEE360 pKa = 3.97FNPSQTSPATAEE372 pKa = 3.97PVRR375 pKa = 11.84DD376 pKa = 3.59EE377 pKa = 4.32VSIEE381 pKa = 3.68TDD383 pKa = 3.46IIDD386 pKa = 3.84EE387 pKa = 4.46VNN389 pKa = 2.86
MM1 pKa = 7.31ATVKK5 pKa = 9.76WADD8 pKa = 2.98ASEE11 pKa = 4.15PQRR14 pKa = 11.84GRR16 pKa = 11.84QGRR19 pKa = 11.84IPYY22 pKa = 9.97SLYY25 pKa = 10.99SPLLVDD31 pKa = 5.73SEE33 pKa = 4.75QPWKK37 pKa = 10.7VIPRR41 pKa = 11.84NLVPINKK48 pKa = 8.93KK49 pKa = 10.4DD50 pKa = 3.45KK51 pKa = 11.0NKK53 pKa = 10.73LIGYY57 pKa = 7.6WNVQKK62 pKa = 10.81RR63 pKa = 11.84FRR65 pKa = 11.84TRR67 pKa = 11.84KK68 pKa = 8.8GKK70 pKa = 10.37RR71 pKa = 11.84VDD73 pKa = 4.23LSPKK77 pKa = 9.28LHH79 pKa = 6.82FYY81 pKa = 11.19YY82 pKa = 10.7LGTGPHH88 pKa = 6.76KK89 pKa = 10.21DD90 pKa = 2.84AKK92 pKa = 10.2FRR94 pKa = 11.84EE95 pKa = 4.43RR96 pKa = 11.84VEE98 pKa = 4.13GVVWVAVDD106 pKa = 3.79GAKK109 pKa = 9.47TEE111 pKa = 3.99PTGYY115 pKa = 10.26GVRR118 pKa = 11.84RR119 pKa = 11.84KK120 pKa = 10.01NSEE123 pKa = 3.64PEE125 pKa = 3.74IPHH128 pKa = 6.81FNQKK132 pKa = 9.87LPNGVTVVEE141 pKa = 4.8EE142 pKa = 4.2PDD144 pKa = 3.07SRR146 pKa = 11.84APSRR150 pKa = 11.84SQSRR154 pKa = 11.84SQSRR158 pKa = 11.84GRR160 pKa = 11.84GEE162 pKa = 4.47SKK164 pKa = 9.89PQSRR168 pKa = 11.84NPSSDD173 pKa = 3.46RR174 pKa = 11.84NHH176 pKa = 6.45NSQDD180 pKa = 3.98DD181 pKa = 3.55IMKK184 pKa = 10.59AVAAALKK191 pKa = 10.67SLGFDD196 pKa = 3.67KK197 pKa = 10.89PQEE200 pKa = 4.05KK201 pKa = 10.13DD202 pKa = 3.04KK203 pKa = 11.44KK204 pKa = 9.11SAKK207 pKa = 9.64TGTPKK212 pKa = 10.47PSRR215 pKa = 11.84NQSPASSQTSAKK227 pKa = 10.2SLARR231 pKa = 11.84SQSSEE236 pKa = 3.88TKK238 pKa = 7.93EE239 pKa = 4.32QKK241 pKa = 10.46HH242 pKa = 5.47EE243 pKa = 4.05MQKK246 pKa = 10.2PRR248 pKa = 11.84WKK250 pKa = 10.22RR251 pKa = 11.84QPNDD255 pKa = 3.49DD256 pKa = 3.43VTSNVTQCFGPRR268 pKa = 11.84DD269 pKa = 3.6LDD271 pKa = 3.95HH272 pKa = 7.0NFGSAGVVANGVKK285 pKa = 10.53AKK287 pKa = 10.29GYY289 pKa = 8.25PQFAEE294 pKa = 4.56LVPSTAAMLFDD305 pKa = 3.29SHH307 pKa = 7.6IVSKK311 pKa = 10.89EE312 pKa = 3.77SGNTVVLTFTTRR324 pKa = 11.84VTVPKK329 pKa = 10.31DD330 pKa = 3.33HH331 pKa = 6.76PHH333 pKa = 6.16LGKK336 pKa = 10.32FLEE339 pKa = 4.34EE340 pKa = 3.99LNAFTRR346 pKa = 11.84EE347 pKa = 4.19MQQHH351 pKa = 6.7PLLNPSALEE360 pKa = 3.97FNPSQTSPATAEE372 pKa = 3.97PVRR375 pKa = 11.84DD376 pKa = 3.59EE377 pKa = 4.32VSIEE381 pKa = 3.68TDD383 pKa = 3.46IIDD386 pKa = 3.84EE387 pKa = 4.46VNN389 pKa = 2.86
Molecular weight: 43.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12928 |
77 |
6758 |
1616.0 |
180.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.799 ± 0.205 | 3.349 ± 0.316 |
5.399 ± 0.411 | 4.223 ± 0.335 |
5.832 ± 0.2 | 6.443 ± 0.16 |
1.756 ± 0.143 | 5.268 ± 0.196 |
6.002 ± 0.602 | 8.609 ± 0.292 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.242 | 5.716 ± 0.227 |
3.233 ± 0.316 | 2.924 ± 0.325 |
3.311 ± 0.264 | 7.008 ± 0.505 |
6.026 ± 0.477 | 10.164 ± 0.225 |
1.392 ± 0.185 | 4.409 ± 0.202 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
