
Cellulophaga phage phi12:3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Helsingorvirus; Cellulophaga virus Cba121; Cellulophaga phage phi12:1
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
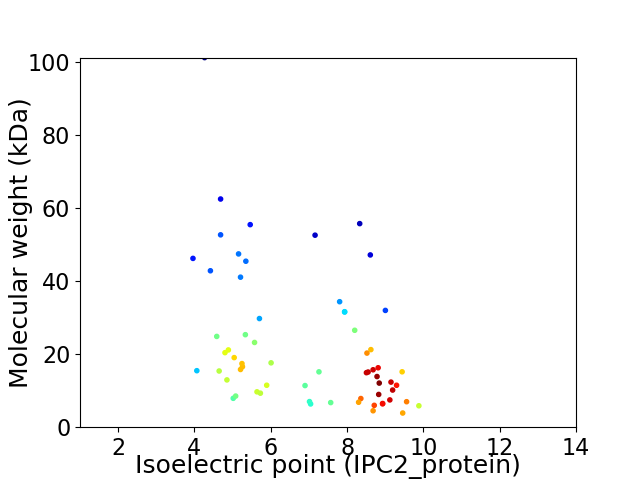
Virtual 2D-PAGE plot for 64 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9ZY40|R9ZY40_9CAUD N-acetylmuramoyl-L-alanine amidase OS=Cellulophaga phage phi12:3 OX=1327979 GN=Phi12:3_gp63 PE=4 SV=1
MM1 pKa = 7.7AYY3 pKa = 9.42IEE5 pKa = 4.88KK6 pKa = 10.32YY7 pKa = 8.06YY8 pKa = 11.0HH9 pKa = 6.52NYY11 pKa = 9.48CDD13 pKa = 4.92SFGTACKK20 pKa = 10.56VSILEE25 pKa = 3.88RR26 pKa = 11.84DD27 pKa = 3.82YY28 pKa = 11.38EE29 pKa = 4.46GSAIEE34 pKa = 5.48LEE36 pKa = 4.38AQFPPFKK43 pKa = 9.76KK44 pKa = 9.23TYY46 pKa = 10.67DD47 pKa = 3.41SDD49 pKa = 4.29SDD51 pKa = 4.0FKK53 pKa = 11.37FEE55 pKa = 5.35PIRR58 pKa = 11.84STSGEE63 pKa = 3.92FNIVFGTGNGIDD75 pKa = 3.8LSDD78 pKa = 4.29IEE80 pKa = 4.59TADD83 pKa = 3.23EE84 pKa = 4.0RR85 pKa = 11.84QYY87 pKa = 12.32KK88 pKa = 8.56MIHH91 pKa = 5.61YY92 pKa = 10.39VDD94 pKa = 5.44GSVDD98 pKa = 3.2WIGYY102 pKa = 6.7VVPDD106 pKa = 4.36GFSHH110 pKa = 6.42EE111 pKa = 4.3LRR113 pKa = 11.84GGIYY117 pKa = 10.11FGSLIATDD125 pKa = 4.38GLKK128 pKa = 10.0QLQSLPFVDD137 pKa = 4.77ISGDD141 pKa = 3.47NYY143 pKa = 11.0GLQDD147 pKa = 3.33LTYY150 pKa = 9.27NNGYY154 pKa = 9.15EE155 pKa = 4.37FPFSLVLTEE164 pKa = 4.48ILRR167 pKa = 11.84KK168 pKa = 9.71LDD170 pKa = 3.77LDD172 pKa = 4.49LNTWTAVDD180 pKa = 4.03VYY182 pKa = 9.62EE183 pKa = 5.82KK184 pKa = 11.49NMTQLGNSRR193 pKa = 11.84DD194 pKa = 4.0SDD196 pKa = 4.3PLSQAYY202 pKa = 10.27VNVKK206 pKa = 9.55TYY208 pKa = 10.92INDD211 pKa = 3.56TTRR214 pKa = 11.84KK215 pKa = 10.04DD216 pKa = 3.07IPYY219 pKa = 9.32WADD222 pKa = 2.85AQEE225 pKa = 4.08AMNCFEE231 pKa = 5.04VLNNMCTLFGARR243 pKa = 11.84VSQNKK248 pKa = 9.11GVWTFKK254 pKa = 10.45RR255 pKa = 11.84VNIDD259 pKa = 3.1ANYY262 pKa = 10.74GSGDD266 pKa = 3.46TQRR269 pKa = 11.84YY270 pKa = 3.85WRR272 pKa = 11.84KK273 pKa = 9.08YY274 pKa = 7.57NTLSVYY280 pKa = 10.79LGRR283 pKa = 11.84EE284 pKa = 4.46AINDD288 pKa = 3.89TVNIPCSDD296 pKa = 3.25IDD298 pKa = 3.77EE299 pKa = 4.4VLIGTDD305 pKa = 3.53HH306 pKa = 6.8QISISDD312 pKa = 3.29VYY314 pKa = 10.65AAYY317 pKa = 10.75RR318 pKa = 11.84MNYY321 pKa = 8.79KK322 pKa = 10.04FQLVRR327 pKa = 11.84EE328 pKa = 4.28GDD330 pKa = 3.65TPLNLLTNGNFSVFNNTSPFAAPQDD355 pKa = 3.26WFLWSEE361 pKa = 4.01GTRR364 pKa = 11.84WVPRR368 pKa = 11.84ISPVTLSPTDD378 pKa = 3.27AGGITTAVEE387 pKa = 4.72FGTLTPGLSTNNTDD401 pKa = 3.71PVGQVWASLRR411 pKa = 11.84YY412 pKa = 9.62YY413 pKa = 10.17EE414 pKa = 4.36NKK416 pKa = 10.15SVNEE420 pKa = 3.55GDD422 pKa = 3.84ALYY425 pKa = 8.37FTAWAKK431 pKa = 10.77FGFNNATNPEE441 pKa = 3.88NVYY444 pKa = 11.02APFLRR449 pKa = 11.84CVIVGASGKK458 pKa = 10.22KK459 pKa = 10.03YY460 pKa = 10.57YY461 pKa = 10.3LANNLSDD468 pKa = 5.85EE469 pKa = 4.06PLKK472 pKa = 10.47WINNDD477 pKa = 4.01DD478 pKa = 3.78FDD480 pKa = 4.31TAKK483 pKa = 11.0DD484 pKa = 3.53DD485 pKa = 4.13LFFVLLYY492 pKa = 10.23GFAVGAPEE500 pKa = 4.18SSDD503 pKa = 2.84ISSEE507 pKa = 3.81FGVYY511 pKa = 8.04GWKK514 pKa = 10.26EE515 pKa = 3.62FALTLPEE522 pKa = 4.22IPEE525 pKa = 4.76DD526 pKa = 3.67GLLTFDD532 pKa = 3.37VHH534 pKa = 7.53GLAKK538 pKa = 10.6FEE540 pKa = 4.33GKK542 pKa = 8.45ATNSFAKK549 pKa = 10.2IKK551 pKa = 10.87LNEE554 pKa = 3.94QKK556 pKa = 10.72SNKK559 pKa = 8.9FIYY562 pKa = 9.23WYY564 pKa = 9.32PVRR567 pKa = 11.84EE568 pKa = 4.09GNYY571 pKa = 9.77DD572 pKa = 3.44NGVGRR577 pKa = 11.84LQITGLSLGTIPDD590 pKa = 3.86PNEE593 pKa = 3.41NAEE596 pKa = 4.13AQDD599 pKa = 3.54YY600 pKa = 11.01VYY602 pKa = 11.45YY603 pKa = 10.34NQNKK607 pKa = 9.4NYY609 pKa = 9.67SLEE612 pKa = 4.06VDD614 pKa = 3.52PVEE617 pKa = 4.47VLNGDD622 pKa = 3.72ILDD625 pKa = 3.82QNHH628 pKa = 5.65VSRR631 pKa = 11.84IIVPSNTTGNKK642 pKa = 9.68NFWDD646 pKa = 4.61TIDD649 pKa = 3.63NKK651 pKa = 11.1YY652 pKa = 10.93GGASLGLITVKK663 pKa = 10.59SIMNLYY669 pKa = 9.65FKK671 pKa = 10.42PFQLLEE677 pKa = 4.6GDD679 pKa = 4.38LKK681 pKa = 11.37SQFGNADD688 pKa = 3.18TVYY691 pKa = 9.66TFEE694 pKa = 5.67AIPGKK699 pKa = 9.21SFCLLRR705 pKa = 11.84GTFIQKK711 pKa = 9.98NNYY714 pKa = 8.86IEE716 pKa = 4.41GATFFEE722 pKa = 4.8ITSEE726 pKa = 4.56DD727 pKa = 3.39IPAGGTEE734 pKa = 4.59GGNSLDD740 pKa = 3.55AEE742 pKa = 4.5YY743 pKa = 11.33VLTGRR748 pKa = 11.84KK749 pKa = 8.99RR750 pKa = 11.84CQKK753 pKa = 10.16VDD755 pKa = 3.44NLNTGLAEE763 pKa = 4.46VEE765 pKa = 4.03LDD767 pKa = 3.54DD768 pKa = 5.54VNINSEE774 pKa = 4.6TYY776 pKa = 10.37GQTIWEE782 pKa = 4.31TSAVLGLTLCPIGEE796 pKa = 4.07PDD798 pKa = 3.16KK799 pKa = 11.19YY800 pKa = 11.47YY801 pKa = 10.28WGTDD805 pKa = 3.06SAVYY809 pKa = 10.31DD810 pKa = 3.59SSTFDD815 pKa = 2.93SVGFYY820 pKa = 10.99EE821 pKa = 4.5EE822 pKa = 5.22DD823 pKa = 3.5GGDD826 pKa = 3.46EE827 pKa = 4.31VSCEE831 pKa = 3.81FTNPGGEE838 pKa = 3.95YY839 pKa = 10.59VYY841 pKa = 10.45FLHH844 pKa = 6.9LASLGLVEE852 pKa = 5.44SIEE855 pKa = 4.29TEE857 pKa = 3.93AQDD860 pKa = 4.91EE861 pKa = 4.67IISDD865 pKa = 4.07FQYY868 pKa = 10.26LTDD871 pKa = 3.63VTINSYY877 pKa = 10.54LYY879 pKa = 9.95RR880 pKa = 11.84VLRR883 pKa = 11.84QNYY886 pKa = 5.34VTSVFNDD893 pKa = 3.04IQITFKK899 pKa = 11.02FSS901 pKa = 2.78
MM1 pKa = 7.7AYY3 pKa = 9.42IEE5 pKa = 4.88KK6 pKa = 10.32YY7 pKa = 8.06YY8 pKa = 11.0HH9 pKa = 6.52NYY11 pKa = 9.48CDD13 pKa = 4.92SFGTACKK20 pKa = 10.56VSILEE25 pKa = 3.88RR26 pKa = 11.84DD27 pKa = 3.82YY28 pKa = 11.38EE29 pKa = 4.46GSAIEE34 pKa = 5.48LEE36 pKa = 4.38AQFPPFKK43 pKa = 9.76KK44 pKa = 9.23TYY46 pKa = 10.67DD47 pKa = 3.41SDD49 pKa = 4.29SDD51 pKa = 4.0FKK53 pKa = 11.37FEE55 pKa = 5.35PIRR58 pKa = 11.84STSGEE63 pKa = 3.92FNIVFGTGNGIDD75 pKa = 3.8LSDD78 pKa = 4.29IEE80 pKa = 4.59TADD83 pKa = 3.23EE84 pKa = 4.0RR85 pKa = 11.84QYY87 pKa = 12.32KK88 pKa = 8.56MIHH91 pKa = 5.61YY92 pKa = 10.39VDD94 pKa = 5.44GSVDD98 pKa = 3.2WIGYY102 pKa = 6.7VVPDD106 pKa = 4.36GFSHH110 pKa = 6.42EE111 pKa = 4.3LRR113 pKa = 11.84GGIYY117 pKa = 10.11FGSLIATDD125 pKa = 4.38GLKK128 pKa = 10.0QLQSLPFVDD137 pKa = 4.77ISGDD141 pKa = 3.47NYY143 pKa = 11.0GLQDD147 pKa = 3.33LTYY150 pKa = 9.27NNGYY154 pKa = 9.15EE155 pKa = 4.37FPFSLVLTEE164 pKa = 4.48ILRR167 pKa = 11.84KK168 pKa = 9.71LDD170 pKa = 3.77LDD172 pKa = 4.49LNTWTAVDD180 pKa = 4.03VYY182 pKa = 9.62EE183 pKa = 5.82KK184 pKa = 11.49NMTQLGNSRR193 pKa = 11.84DD194 pKa = 4.0SDD196 pKa = 4.3PLSQAYY202 pKa = 10.27VNVKK206 pKa = 9.55TYY208 pKa = 10.92INDD211 pKa = 3.56TTRR214 pKa = 11.84KK215 pKa = 10.04DD216 pKa = 3.07IPYY219 pKa = 9.32WADD222 pKa = 2.85AQEE225 pKa = 4.08AMNCFEE231 pKa = 5.04VLNNMCTLFGARR243 pKa = 11.84VSQNKK248 pKa = 9.11GVWTFKK254 pKa = 10.45RR255 pKa = 11.84VNIDD259 pKa = 3.1ANYY262 pKa = 10.74GSGDD266 pKa = 3.46TQRR269 pKa = 11.84YY270 pKa = 3.85WRR272 pKa = 11.84KK273 pKa = 9.08YY274 pKa = 7.57NTLSVYY280 pKa = 10.79LGRR283 pKa = 11.84EE284 pKa = 4.46AINDD288 pKa = 3.89TVNIPCSDD296 pKa = 3.25IDD298 pKa = 3.77EE299 pKa = 4.4VLIGTDD305 pKa = 3.53HH306 pKa = 6.8QISISDD312 pKa = 3.29VYY314 pKa = 10.65AAYY317 pKa = 10.75RR318 pKa = 11.84MNYY321 pKa = 8.79KK322 pKa = 10.04FQLVRR327 pKa = 11.84EE328 pKa = 4.28GDD330 pKa = 3.65TPLNLLTNGNFSVFNNTSPFAAPQDD355 pKa = 3.26WFLWSEE361 pKa = 4.01GTRR364 pKa = 11.84WVPRR368 pKa = 11.84ISPVTLSPTDD378 pKa = 3.27AGGITTAVEE387 pKa = 4.72FGTLTPGLSTNNTDD401 pKa = 3.71PVGQVWASLRR411 pKa = 11.84YY412 pKa = 9.62YY413 pKa = 10.17EE414 pKa = 4.36NKK416 pKa = 10.15SVNEE420 pKa = 3.55GDD422 pKa = 3.84ALYY425 pKa = 8.37FTAWAKK431 pKa = 10.77FGFNNATNPEE441 pKa = 3.88NVYY444 pKa = 11.02APFLRR449 pKa = 11.84CVIVGASGKK458 pKa = 10.22KK459 pKa = 10.03YY460 pKa = 10.57YY461 pKa = 10.3LANNLSDD468 pKa = 5.85EE469 pKa = 4.06PLKK472 pKa = 10.47WINNDD477 pKa = 4.01DD478 pKa = 3.78FDD480 pKa = 4.31TAKK483 pKa = 11.0DD484 pKa = 3.53DD485 pKa = 4.13LFFVLLYY492 pKa = 10.23GFAVGAPEE500 pKa = 4.18SSDD503 pKa = 2.84ISSEE507 pKa = 3.81FGVYY511 pKa = 8.04GWKK514 pKa = 10.26EE515 pKa = 3.62FALTLPEE522 pKa = 4.22IPEE525 pKa = 4.76DD526 pKa = 3.67GLLTFDD532 pKa = 3.37VHH534 pKa = 7.53GLAKK538 pKa = 10.6FEE540 pKa = 4.33GKK542 pKa = 8.45ATNSFAKK549 pKa = 10.2IKK551 pKa = 10.87LNEE554 pKa = 3.94QKK556 pKa = 10.72SNKK559 pKa = 8.9FIYY562 pKa = 9.23WYY564 pKa = 9.32PVRR567 pKa = 11.84EE568 pKa = 4.09GNYY571 pKa = 9.77DD572 pKa = 3.44NGVGRR577 pKa = 11.84LQITGLSLGTIPDD590 pKa = 3.86PNEE593 pKa = 3.41NAEE596 pKa = 4.13AQDD599 pKa = 3.54YY600 pKa = 11.01VYY602 pKa = 11.45YY603 pKa = 10.34NQNKK607 pKa = 9.4NYY609 pKa = 9.67SLEE612 pKa = 4.06VDD614 pKa = 3.52PVEE617 pKa = 4.47VLNGDD622 pKa = 3.72ILDD625 pKa = 3.82QNHH628 pKa = 5.65VSRR631 pKa = 11.84IIVPSNTTGNKK642 pKa = 9.68NFWDD646 pKa = 4.61TIDD649 pKa = 3.63NKK651 pKa = 11.1YY652 pKa = 10.93GGASLGLITVKK663 pKa = 10.59SIMNLYY669 pKa = 9.65FKK671 pKa = 10.42PFQLLEE677 pKa = 4.6GDD679 pKa = 4.38LKK681 pKa = 11.37SQFGNADD688 pKa = 3.18TVYY691 pKa = 9.66TFEE694 pKa = 5.67AIPGKK699 pKa = 9.21SFCLLRR705 pKa = 11.84GTFIQKK711 pKa = 9.98NNYY714 pKa = 8.86IEE716 pKa = 4.41GATFFEE722 pKa = 4.8ITSEE726 pKa = 4.56DD727 pKa = 3.39IPAGGTEE734 pKa = 4.59GGNSLDD740 pKa = 3.55AEE742 pKa = 4.5YY743 pKa = 11.33VLTGRR748 pKa = 11.84KK749 pKa = 8.99RR750 pKa = 11.84CQKK753 pKa = 10.16VDD755 pKa = 3.44NLNTGLAEE763 pKa = 4.46VEE765 pKa = 4.03LDD767 pKa = 3.54DD768 pKa = 5.54VNINSEE774 pKa = 4.6TYY776 pKa = 10.37GQTIWEE782 pKa = 4.31TSAVLGLTLCPIGEE796 pKa = 4.07PDD798 pKa = 3.16KK799 pKa = 11.19YY800 pKa = 11.47YY801 pKa = 10.28WGTDD805 pKa = 3.06SAVYY809 pKa = 10.31DD810 pKa = 3.59SSTFDD815 pKa = 2.93SVGFYY820 pKa = 10.99EE821 pKa = 4.5EE822 pKa = 5.22DD823 pKa = 3.5GGDD826 pKa = 3.46EE827 pKa = 4.31VSCEE831 pKa = 3.81FTNPGGEE838 pKa = 3.95YY839 pKa = 10.59VYY841 pKa = 10.45FLHH844 pKa = 6.9LASLGLVEE852 pKa = 5.44SIEE855 pKa = 4.29TEE857 pKa = 3.93AQDD860 pKa = 4.91EE861 pKa = 4.67IISDD865 pKa = 4.07FQYY868 pKa = 10.26LTDD871 pKa = 3.63VTINSYY877 pKa = 10.54LYY879 pKa = 9.95RR880 pKa = 11.84VLRR883 pKa = 11.84QNYY886 pKa = 5.34VTSVFNDD893 pKa = 3.04IQITFKK899 pKa = 11.02FSS901 pKa = 2.78
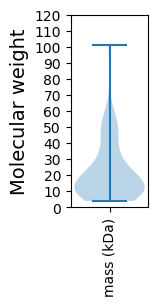
Molecular weight: 101.22 kDa
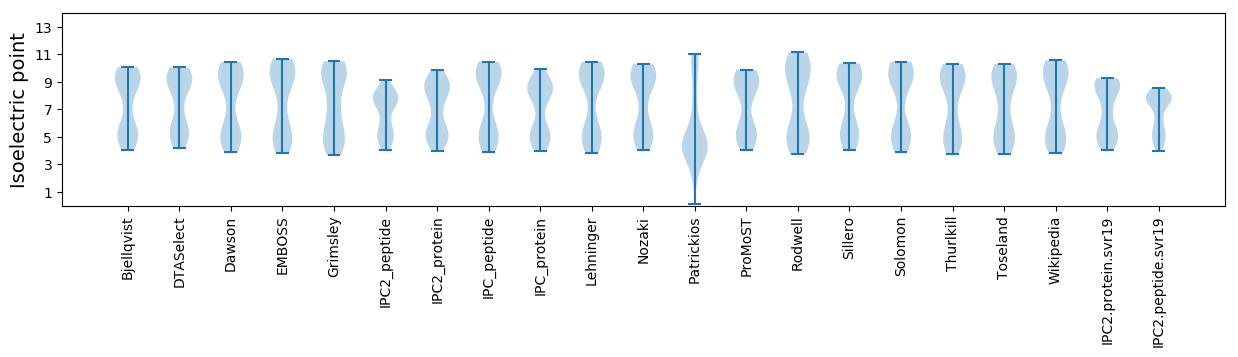
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S0A0Y5|S0A0Y5_9CAUD VirE N-domain protein OS=Cellulophaga phage phi12:3 OX=1327979 GN=Phi12:3_gp21 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.21ISEE5 pKa = 4.33TKK7 pKa = 10.05KK8 pKa = 10.49RR9 pKa = 11.84EE10 pKa = 4.0GVYY13 pKa = 10.45CCAYY17 pKa = 9.07GCKK20 pKa = 9.94GHH22 pKa = 7.02PEE24 pKa = 3.77QRR26 pKa = 11.84KK27 pKa = 9.5GMLCHH32 pKa = 6.11KK33 pKa = 10.03HH34 pKa = 4.72YY35 pKa = 10.97ARR37 pKa = 11.84LMRR40 pKa = 11.84EE41 pKa = 3.98RR42 pKa = 11.84SPKK45 pKa = 9.29KK46 pKa = 10.32VRR48 pKa = 11.84YY49 pKa = 9.11SQAKK53 pKa = 8.88QKK55 pKa = 10.77AKK57 pKa = 10.7SRR59 pKa = 11.84GIDD62 pKa = 3.47FTITLEE68 pKa = 3.88WFLRR72 pKa = 11.84FCDD75 pKa = 3.25RR76 pKa = 11.84TGYY79 pKa = 8.75MSKK82 pKa = 10.47GRR84 pKa = 11.84RR85 pKa = 11.84GQNATLDD92 pKa = 3.82RR93 pKa = 11.84RR94 pKa = 11.84CNLHH98 pKa = 7.13GYY100 pKa = 9.53HH101 pKa = 6.3SWNIQILTNRR111 pKa = 11.84QNASKK116 pKa = 10.71GNRR119 pKa = 11.84PSGEE123 pKa = 4.23HH124 pKa = 6.38FDD126 pKa = 4.52CPFF129 pKa = 4.35
MM1 pKa = 7.59KK2 pKa = 10.21ISEE5 pKa = 4.33TKK7 pKa = 10.05KK8 pKa = 10.49RR9 pKa = 11.84EE10 pKa = 4.0GVYY13 pKa = 10.45CCAYY17 pKa = 9.07GCKK20 pKa = 9.94GHH22 pKa = 7.02PEE24 pKa = 3.77QRR26 pKa = 11.84KK27 pKa = 9.5GMLCHH32 pKa = 6.11KK33 pKa = 10.03HH34 pKa = 4.72YY35 pKa = 10.97ARR37 pKa = 11.84LMRR40 pKa = 11.84EE41 pKa = 3.98RR42 pKa = 11.84SPKK45 pKa = 9.29KK46 pKa = 10.32VRR48 pKa = 11.84YY49 pKa = 9.11SQAKK53 pKa = 8.88QKK55 pKa = 10.77AKK57 pKa = 10.7SRR59 pKa = 11.84GIDD62 pKa = 3.47FTITLEE68 pKa = 3.88WFLRR72 pKa = 11.84FCDD75 pKa = 3.25RR76 pKa = 11.84TGYY79 pKa = 8.75MSKK82 pKa = 10.47GRR84 pKa = 11.84RR85 pKa = 11.84GQNATLDD92 pKa = 3.82RR93 pKa = 11.84RR94 pKa = 11.84CNLHH98 pKa = 7.13GYY100 pKa = 9.53HH101 pKa = 6.3SWNIQILTNRR111 pKa = 11.84QNASKK116 pKa = 10.71GNRR119 pKa = 11.84PSGEE123 pKa = 4.23HH124 pKa = 6.38FDD126 pKa = 4.52CPFF129 pKa = 4.35
Molecular weight: 15.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12570 |
33 |
901 |
196.4 |
22.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.165 ± 0.47 | 1.082 ± 0.127 |
6.667 ± 0.21 | 6.547 ± 0.274 |
4.678 ± 0.221 | 6.364 ± 0.426 |
1.329 ± 0.156 | 7.804 ± 0.283 |
8.958 ± 0.61 | 8.234 ± 0.26 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.578 ± 0.227 | 5.831 ± 0.286 |
3.333 ± 0.211 | 3.532 ± 0.213 |
3.723 ± 0.221 | 6.929 ± 0.326 |
5.306 ± 0.364 | 5.442 ± 0.199 |
1.392 ± 0.123 | 4.105 ± 0.281 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
