
Sanxia water strider virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
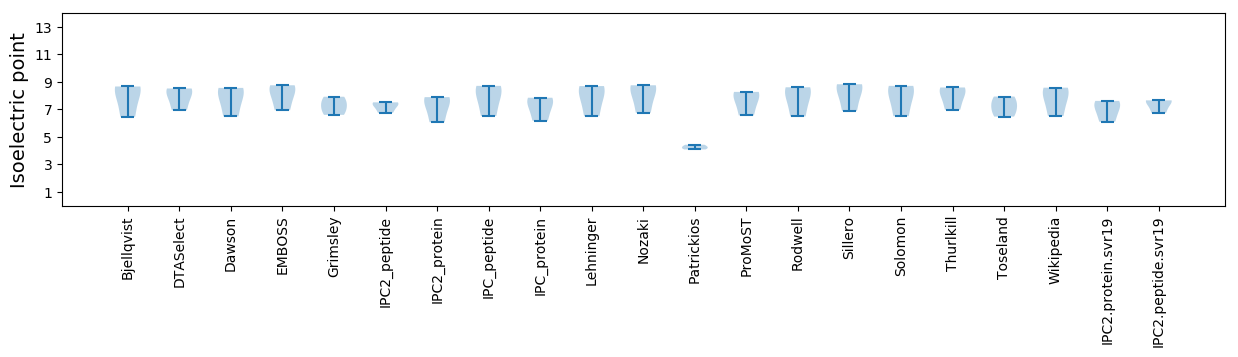
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEV2|A0A1L3KEV2_9VIRU Uncharacterized protein OS=Sanxia water strider virus 10 OX=1923394 PE=4 SV=1
MM1 pKa = 7.66QEE3 pKa = 4.12LLSGKK8 pKa = 9.29EE9 pKa = 3.92VADD12 pKa = 5.18DD13 pKa = 2.94IHH15 pKa = 8.33VFVKK19 pKa = 10.12QEE21 pKa = 3.53PHH23 pKa = 6.1KK24 pKa = 10.39RR25 pKa = 11.84AKK27 pKa = 10.36LDD29 pKa = 3.22EE30 pKa = 4.11GRR32 pKa = 11.84YY33 pKa = 9.24RR34 pKa = 11.84LISAVSAVDD43 pKa = 3.41TMVDD47 pKa = 3.84RR48 pKa = 11.84ILFGPLQAKK57 pKa = 9.44VLEE60 pKa = 4.79TVGQTPTMIGWTPLQGGYY78 pKa = 8.4RR79 pKa = 11.84HH80 pKa = 6.13FRR82 pKa = 11.84RR83 pKa = 11.84MLAKK87 pKa = 9.87RR88 pKa = 11.84VMCADD93 pKa = 3.3KK94 pKa = 11.14SSWDD98 pKa = 3.22WTVPGWLVDD107 pKa = 4.02FWEE110 pKa = 4.8DD111 pKa = 3.83FVLSLCVDD119 pKa = 3.45APEE122 pKa = 3.47WWQRR126 pKa = 11.84LVRR129 pKa = 11.84MRR131 pKa = 11.84FRR133 pKa = 11.84MLFSEE138 pKa = 5.22AVFQFADD145 pKa = 3.93GTRR148 pKa = 11.84VKK150 pKa = 10.74QPVKK154 pKa = 10.55GVMKK158 pKa = 10.28SGCLLTILLNSISQVIIHH176 pKa = 6.44VIVQLLLGNDD186 pKa = 3.46PMEE189 pKa = 4.29GLPYY193 pKa = 10.64NIGDD197 pKa = 3.89DD198 pKa = 3.61TTQAFLEE205 pKa = 4.25YY206 pKa = 9.83FRR208 pKa = 11.84EE209 pKa = 4.11YY210 pKa = 10.16FAEE213 pKa = 4.01IEE215 pKa = 4.12KK216 pKa = 10.89LGIRR220 pKa = 11.84VKK222 pKa = 9.89EE223 pKa = 4.04VKK225 pKa = 10.1IQDD228 pKa = 3.25WVEE231 pKa = 3.55FAGFYY236 pKa = 10.16IGCSEE241 pKa = 4.12TWPVYY246 pKa = 8.65WEE248 pKa = 3.69KK249 pKa = 10.83HH250 pKa = 5.54LYY252 pKa = 9.71SVAHH256 pKa = 6.11QEE258 pKa = 5.09DD259 pKa = 3.9KK260 pKa = 11.22LLPQTLFMYY269 pKa = 10.65QMLYY273 pKa = 10.38AAQPSMLSFIRR284 pKa = 11.84KK285 pKa = 8.48HH286 pKa = 6.1LKK288 pKa = 10.08EE289 pKa = 4.81LDD291 pKa = 3.5PTLCKK296 pKa = 9.99PLWVLRR302 pKa = 11.84GYY304 pKa = 11.28VDD306 pKa = 3.17GTLPLHH312 pKa = 6.25ITSNWII318 pKa = 3.15
MM1 pKa = 7.66QEE3 pKa = 4.12LLSGKK8 pKa = 9.29EE9 pKa = 3.92VADD12 pKa = 5.18DD13 pKa = 2.94IHH15 pKa = 8.33VFVKK19 pKa = 10.12QEE21 pKa = 3.53PHH23 pKa = 6.1KK24 pKa = 10.39RR25 pKa = 11.84AKK27 pKa = 10.36LDD29 pKa = 3.22EE30 pKa = 4.11GRR32 pKa = 11.84YY33 pKa = 9.24RR34 pKa = 11.84LISAVSAVDD43 pKa = 3.41TMVDD47 pKa = 3.84RR48 pKa = 11.84ILFGPLQAKK57 pKa = 9.44VLEE60 pKa = 4.79TVGQTPTMIGWTPLQGGYY78 pKa = 8.4RR79 pKa = 11.84HH80 pKa = 6.13FRR82 pKa = 11.84RR83 pKa = 11.84MLAKK87 pKa = 9.87RR88 pKa = 11.84VMCADD93 pKa = 3.3KK94 pKa = 11.14SSWDD98 pKa = 3.22WTVPGWLVDD107 pKa = 4.02FWEE110 pKa = 4.8DD111 pKa = 3.83FVLSLCVDD119 pKa = 3.45APEE122 pKa = 3.47WWQRR126 pKa = 11.84LVRR129 pKa = 11.84MRR131 pKa = 11.84FRR133 pKa = 11.84MLFSEE138 pKa = 5.22AVFQFADD145 pKa = 3.93GTRR148 pKa = 11.84VKK150 pKa = 10.74QPVKK154 pKa = 10.55GVMKK158 pKa = 10.28SGCLLTILLNSISQVIIHH176 pKa = 6.44VIVQLLLGNDD186 pKa = 3.46PMEE189 pKa = 4.29GLPYY193 pKa = 10.64NIGDD197 pKa = 3.89DD198 pKa = 3.61TTQAFLEE205 pKa = 4.25YY206 pKa = 9.83FRR208 pKa = 11.84EE209 pKa = 4.11YY210 pKa = 10.16FAEE213 pKa = 4.01IEE215 pKa = 4.12KK216 pKa = 10.89LGIRR220 pKa = 11.84VKK222 pKa = 9.89EE223 pKa = 4.04VKK225 pKa = 10.1IQDD228 pKa = 3.25WVEE231 pKa = 3.55FAGFYY236 pKa = 10.16IGCSEE241 pKa = 4.12TWPVYY246 pKa = 8.65WEE248 pKa = 3.69KK249 pKa = 10.83HH250 pKa = 5.54LYY252 pKa = 9.71SVAHH256 pKa = 6.11QEE258 pKa = 5.09DD259 pKa = 3.9KK260 pKa = 11.22LLPQTLFMYY269 pKa = 10.65QMLYY273 pKa = 10.38AAQPSMLSFIRR284 pKa = 11.84KK285 pKa = 8.48HH286 pKa = 6.1LKK288 pKa = 10.08EE289 pKa = 4.81LDD291 pKa = 3.5PTLCKK296 pKa = 9.99PLWVLRR302 pKa = 11.84GYY304 pKa = 11.28VDD306 pKa = 3.17GTLPLHH312 pKa = 6.25ITSNWII318 pKa = 3.15
Molecular weight: 36.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEU6|A0A1L3KEU6_9VIRU RNA-directed RNA polymerase OS=Sanxia water strider virus 10 OX=1923394 PE=4 SV=1
MM1 pKa = 7.59PPKK4 pKa = 10.12SVNPIKK10 pKa = 10.57EE11 pKa = 4.35RR12 pKa = 11.84ICPYY16 pKa = 10.07CGSKK20 pKa = 10.84FMGKK24 pKa = 8.98RR25 pKa = 11.84AMMTHH30 pKa = 7.48LLSAHH35 pKa = 6.81PGSSQTKK42 pKa = 9.41NRR44 pKa = 11.84AQAAPKK50 pKa = 8.4PQRR53 pKa = 11.84VKK55 pKa = 11.09AKK57 pKa = 9.86GPSGSNVARR66 pKa = 11.84VASGRR71 pKa = 11.84DD72 pKa = 3.15IIATVSVASTKK83 pKa = 10.22SVGYY87 pKa = 10.08NVVEE91 pKa = 4.19LFVNPRR97 pKa = 11.84LLPSTRR103 pKa = 11.84LCQEE107 pKa = 3.6SKK109 pKa = 8.9MWSRR113 pKa = 11.84WLPVTLMVEE122 pKa = 4.26IRR124 pKa = 11.84TSSPKK129 pKa = 10.18IIGGMYY135 pKa = 8.74IAAWTPDD142 pKa = 3.26SKK144 pKa = 11.72EE145 pKa = 4.02NLPSGADD152 pKa = 3.19AVRR155 pKa = 11.84RR156 pKa = 11.84LATWPRR162 pKa = 11.84SLEE165 pKa = 3.97RR166 pKa = 11.84HH167 pKa = 5.53VSTDD171 pKa = 2.85SQFQIPVDD179 pKa = 4.31ASRR182 pKa = 11.84KK183 pKa = 8.75WYY185 pKa = 8.97TFDD188 pKa = 5.95SGDD191 pKa = 3.58QTDD194 pKa = 3.77TTHH197 pKa = 7.55GIFVLLLSAPISGLTGDD214 pKa = 3.81IQFSVHH220 pKa = 5.63LHH222 pKa = 4.0WTIKK226 pKa = 10.42FDD228 pKa = 4.01GPEE231 pKa = 4.15LEE233 pKa = 4.52MSGPQPGDD241 pKa = 3.64FIYY244 pKa = 10.83ADD246 pKa = 3.65SGYY249 pKa = 8.79TPYY252 pKa = 10.71FSDD255 pKa = 5.34SSNDD259 pKa = 2.84WWGGKK264 pKa = 8.55YY265 pKa = 10.43LSLKK269 pKa = 8.18HH270 pKa = 5.89TNGGSMVPFPTAKK283 pKa = 9.86HH284 pKa = 5.5DD285 pKa = 3.85VVYY288 pKa = 10.78KK289 pKa = 8.3CTEE292 pKa = 4.31PIGYY296 pKa = 9.21YY297 pKa = 10.07KK298 pKa = 11.11SNGTLHH304 pKa = 5.92NASYY308 pKa = 11.13GVLAKK313 pKa = 10.66NYY315 pKa = 8.01TSYY318 pKa = 11.62NIMALFVSKK327 pKa = 10.61EE328 pKa = 3.82DD329 pKa = 3.13AFKK332 pKa = 10.57YY333 pKa = 10.36VKK335 pKa = 10.69DD336 pKa = 3.7GGEE339 pKa = 3.85TGLLQFYY346 pKa = 10.99SDD348 pKa = 4.89GPWVSPPNPAWEE360 pKa = 4.28TVPVPPTTIEE370 pKa = 4.04VPGFTNRR377 pKa = 11.84RR378 pKa = 11.84IALDD382 pKa = 3.39RR383 pKa = 11.84VDD385 pKa = 5.45RR386 pKa = 11.84DD387 pKa = 3.28ILLEE391 pKa = 3.99TLFQRR396 pKa = 11.84ISSLEE401 pKa = 3.59EE402 pKa = 4.05RR403 pKa = 11.84IRR405 pKa = 11.84SLQVTPASSTSSSMIVLHH423 pKa = 6.17EE424 pKa = 4.29PPVEE428 pKa = 4.01EE429 pKa = 4.44EE430 pKa = 4.26SLNRR434 pKa = 11.84PPGPDD439 pKa = 2.87SS440 pKa = 3.52
MM1 pKa = 7.59PPKK4 pKa = 10.12SVNPIKK10 pKa = 10.57EE11 pKa = 4.35RR12 pKa = 11.84ICPYY16 pKa = 10.07CGSKK20 pKa = 10.84FMGKK24 pKa = 8.98RR25 pKa = 11.84AMMTHH30 pKa = 7.48LLSAHH35 pKa = 6.81PGSSQTKK42 pKa = 9.41NRR44 pKa = 11.84AQAAPKK50 pKa = 8.4PQRR53 pKa = 11.84VKK55 pKa = 11.09AKK57 pKa = 9.86GPSGSNVARR66 pKa = 11.84VASGRR71 pKa = 11.84DD72 pKa = 3.15IIATVSVASTKK83 pKa = 10.22SVGYY87 pKa = 10.08NVVEE91 pKa = 4.19LFVNPRR97 pKa = 11.84LLPSTRR103 pKa = 11.84LCQEE107 pKa = 3.6SKK109 pKa = 8.9MWSRR113 pKa = 11.84WLPVTLMVEE122 pKa = 4.26IRR124 pKa = 11.84TSSPKK129 pKa = 10.18IIGGMYY135 pKa = 8.74IAAWTPDD142 pKa = 3.26SKK144 pKa = 11.72EE145 pKa = 4.02NLPSGADD152 pKa = 3.19AVRR155 pKa = 11.84RR156 pKa = 11.84LATWPRR162 pKa = 11.84SLEE165 pKa = 3.97RR166 pKa = 11.84HH167 pKa = 5.53VSTDD171 pKa = 2.85SQFQIPVDD179 pKa = 4.31ASRR182 pKa = 11.84KK183 pKa = 8.75WYY185 pKa = 8.97TFDD188 pKa = 5.95SGDD191 pKa = 3.58QTDD194 pKa = 3.77TTHH197 pKa = 7.55GIFVLLLSAPISGLTGDD214 pKa = 3.81IQFSVHH220 pKa = 5.63LHH222 pKa = 4.0WTIKK226 pKa = 10.42FDD228 pKa = 4.01GPEE231 pKa = 4.15LEE233 pKa = 4.52MSGPQPGDD241 pKa = 3.64FIYY244 pKa = 10.83ADD246 pKa = 3.65SGYY249 pKa = 8.79TPYY252 pKa = 10.71FSDD255 pKa = 5.34SSNDD259 pKa = 2.84WWGGKK264 pKa = 8.55YY265 pKa = 10.43LSLKK269 pKa = 8.18HH270 pKa = 5.89TNGGSMVPFPTAKK283 pKa = 9.86HH284 pKa = 5.5DD285 pKa = 3.85VVYY288 pKa = 10.78KK289 pKa = 8.3CTEE292 pKa = 4.31PIGYY296 pKa = 9.21YY297 pKa = 10.07KK298 pKa = 11.11SNGTLHH304 pKa = 5.92NASYY308 pKa = 11.13GVLAKK313 pKa = 10.66NYY315 pKa = 8.01TSYY318 pKa = 11.62NIMALFVSKK327 pKa = 10.61EE328 pKa = 3.82DD329 pKa = 3.13AFKK332 pKa = 10.57YY333 pKa = 10.36VKK335 pKa = 10.69DD336 pKa = 3.7GGEE339 pKa = 3.85TGLLQFYY346 pKa = 10.99SDD348 pKa = 4.89GPWVSPPNPAWEE360 pKa = 4.28TVPVPPTTIEE370 pKa = 4.04VPGFTNRR377 pKa = 11.84RR378 pKa = 11.84IALDD382 pKa = 3.39RR383 pKa = 11.84VDD385 pKa = 5.45RR386 pKa = 11.84DD387 pKa = 3.28ILLEE391 pKa = 3.99TLFQRR396 pKa = 11.84ISSLEE401 pKa = 3.59EE402 pKa = 4.05RR403 pKa = 11.84IRR405 pKa = 11.84SLQVTPASSTSSSMIVLHH423 pKa = 6.17EE424 pKa = 4.29PPVEE428 pKa = 4.01EE429 pKa = 4.44EE430 pKa = 4.26SLNRR434 pKa = 11.84PPGPDD439 pKa = 2.87SS440 pKa = 3.52
Molecular weight: 48.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1328 |
318 |
570 |
442.7 |
49.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.702 ± 0.8 | 1.205 ± 0.135 |
5.045 ± 0.199 | 4.819 ± 0.351 |
4.292 ± 0.365 | 6.25 ± 0.332 |
2.033 ± 0.235 | 4.593 ± 0.378 |
5.271 ± 0.124 | 9.639 ± 0.978 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.937 ± 0.26 | 2.56 ± 0.432 |
6.551 ± 0.866 | 3.389 ± 0.495 |
5.045 ± 0.158 | 8.208 ± 1.316 |
6.401 ± 0.51 | 8.283 ± 0.511 |
2.711 ± 0.32 | 4.066 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
