
Spider associated circular virus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
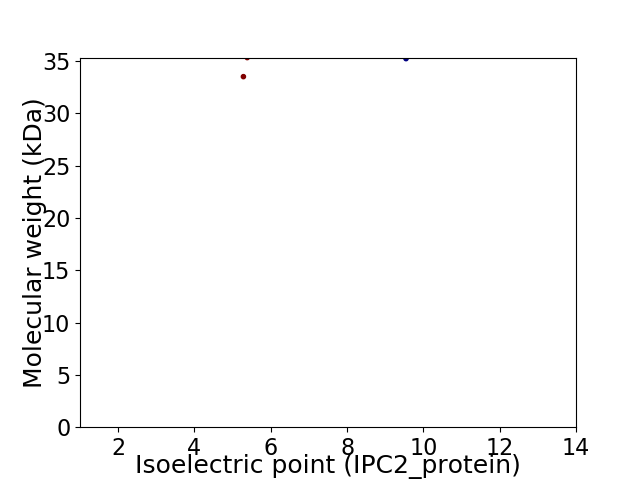
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BP77|A0A346BP77_9VIRU Putative capsid protein OS=Spider associated circular virus 2 OX=2293303 PE=4 SV=1
MM1 pKa = 7.9PFNFQARR8 pKa = 11.84YY9 pKa = 9.97ALLTYY14 pKa = 8.97AQCDD18 pKa = 3.43ALDD21 pKa = 3.81GFRR24 pKa = 11.84VMDD27 pKa = 4.22HH28 pKa = 6.83LSGLGAEE35 pKa = 4.95CIISRR40 pKa = 11.84EE41 pKa = 3.84VHH43 pKa = 5.97ANGGIHH49 pKa = 6.31LHH51 pKa = 6.32CFVDD55 pKa = 4.63FEE57 pKa = 4.87RR58 pKa = 11.84KK59 pKa = 8.77FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84TDD66 pKa = 2.64IFDD69 pKa = 3.14VDD71 pKa = 3.53GRR73 pKa = 11.84HH74 pKa = 5.9PNIVSSYY81 pKa = 7.57GSPWGGYY88 pKa = 10.22DD89 pKa = 3.69YY90 pKa = 10.87AIKK93 pKa = 10.81DD94 pKa = 3.36GDD96 pKa = 4.29VICGGLEE103 pKa = 3.73RR104 pKa = 11.84PEE106 pKa = 4.01EE107 pKa = 4.12PRR109 pKa = 11.84SKK111 pKa = 10.41RR112 pKa = 11.84VAKK115 pKa = 10.18DD116 pKa = 2.9WNAWTEE122 pKa = 3.76ITNARR127 pKa = 11.84DD128 pKa = 3.44RR129 pKa = 11.84SHH131 pKa = 6.61FWEE134 pKa = 5.0LVHH137 pKa = 8.01HH138 pKa = 7.16LDD140 pKa = 4.88PKK142 pKa = 10.84AAACNHH148 pKa = 5.45GQLAKK153 pKa = 9.28YY154 pKa = 10.38AEE156 pKa = 4.04WRR158 pKa = 11.84FRR160 pKa = 11.84DD161 pKa = 3.88TPAKK165 pKa = 9.94YY166 pKa = 9.4EE167 pKa = 4.26SPGGIEE173 pKa = 4.88FIGGDD178 pKa = 3.37VDD180 pKa = 6.43GRR182 pKa = 11.84DD183 pKa = 3.34AWCDD187 pKa = 3.17QSGIRR192 pKa = 11.84SGEE195 pKa = 3.79PLVEE199 pKa = 4.15LKK201 pKa = 10.54KK202 pKa = 10.98ASNDD206 pKa = 3.4DD207 pKa = 3.12VKK209 pKa = 11.37YY210 pKa = 10.88AIFDD214 pKa = 4.73DD215 pKa = 3.41IRR217 pKa = 11.84GGIKK221 pKa = 9.97FFPAFKK227 pKa = 10.19EE228 pKa = 4.03WMGAQAYY235 pKa = 6.96VTVKK239 pKa = 10.19EE240 pKa = 4.63LYY242 pKa = 10.09RR243 pKa = 11.84EE244 pKa = 3.85PALIKK249 pKa = 9.55WGKK252 pKa = 8.34PSIWLANDD260 pKa = 3.26DD261 pKa = 3.48PRR263 pKa = 11.84NYY265 pKa = 10.08VEE267 pKa = 4.09QSDD270 pKa = 4.08RR271 pKa = 11.84SWLEE275 pKa = 3.69EE276 pKa = 3.51NCTFIEE282 pKa = 4.07VTEE285 pKa = 4.81AIFRR289 pKa = 11.84ANTEE293 pKa = 3.84
MM1 pKa = 7.9PFNFQARR8 pKa = 11.84YY9 pKa = 9.97ALLTYY14 pKa = 8.97AQCDD18 pKa = 3.43ALDD21 pKa = 3.81GFRR24 pKa = 11.84VMDD27 pKa = 4.22HH28 pKa = 6.83LSGLGAEE35 pKa = 4.95CIISRR40 pKa = 11.84EE41 pKa = 3.84VHH43 pKa = 5.97ANGGIHH49 pKa = 6.31LHH51 pKa = 6.32CFVDD55 pKa = 4.63FEE57 pKa = 4.87RR58 pKa = 11.84KK59 pKa = 8.77FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84TDD66 pKa = 2.64IFDD69 pKa = 3.14VDD71 pKa = 3.53GRR73 pKa = 11.84HH74 pKa = 5.9PNIVSSYY81 pKa = 7.57GSPWGGYY88 pKa = 10.22DD89 pKa = 3.69YY90 pKa = 10.87AIKK93 pKa = 10.81DD94 pKa = 3.36GDD96 pKa = 4.29VICGGLEE103 pKa = 3.73RR104 pKa = 11.84PEE106 pKa = 4.01EE107 pKa = 4.12PRR109 pKa = 11.84SKK111 pKa = 10.41RR112 pKa = 11.84VAKK115 pKa = 10.18DD116 pKa = 2.9WNAWTEE122 pKa = 3.76ITNARR127 pKa = 11.84DD128 pKa = 3.44RR129 pKa = 11.84SHH131 pKa = 6.61FWEE134 pKa = 5.0LVHH137 pKa = 8.01HH138 pKa = 7.16LDD140 pKa = 4.88PKK142 pKa = 10.84AAACNHH148 pKa = 5.45GQLAKK153 pKa = 9.28YY154 pKa = 10.38AEE156 pKa = 4.04WRR158 pKa = 11.84FRR160 pKa = 11.84DD161 pKa = 3.88TPAKK165 pKa = 9.94YY166 pKa = 9.4EE167 pKa = 4.26SPGGIEE173 pKa = 4.88FIGGDD178 pKa = 3.37VDD180 pKa = 6.43GRR182 pKa = 11.84DD183 pKa = 3.34AWCDD187 pKa = 3.17QSGIRR192 pKa = 11.84SGEE195 pKa = 3.79PLVEE199 pKa = 4.15LKK201 pKa = 10.54KK202 pKa = 10.98ASNDD206 pKa = 3.4DD207 pKa = 3.12VKK209 pKa = 11.37YY210 pKa = 10.88AIFDD214 pKa = 4.73DD215 pKa = 3.41IRR217 pKa = 11.84GGIKK221 pKa = 9.97FFPAFKK227 pKa = 10.19EE228 pKa = 4.03WMGAQAYY235 pKa = 6.96VTVKK239 pKa = 10.19EE240 pKa = 4.63LYY242 pKa = 10.09RR243 pKa = 11.84EE244 pKa = 3.85PALIKK249 pKa = 9.55WGKK252 pKa = 8.34PSIWLANDD260 pKa = 3.26DD261 pKa = 3.48PRR263 pKa = 11.84NYY265 pKa = 10.08VEE267 pKa = 4.09QSDD270 pKa = 4.08RR271 pKa = 11.84SWLEE275 pKa = 3.69EE276 pKa = 3.51NCTFIEE282 pKa = 4.07VTEE285 pKa = 4.81AIFRR289 pKa = 11.84ANTEE293 pKa = 3.84
Molecular weight: 33.51 kDa
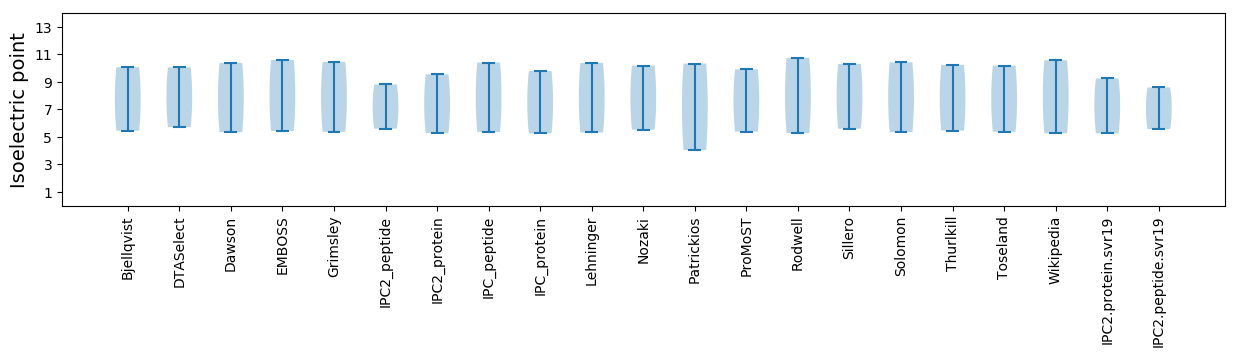
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BP77|A0A346BP77_9VIRU Putative capsid protein OS=Spider associated circular virus 2 OX=2293303 PE=4 SV=1
MM1 pKa = 7.82AYY3 pKa = 10.28ARR5 pKa = 11.84RR6 pKa = 11.84NRR8 pKa = 11.84YY9 pKa = 8.67RR10 pKa = 11.84RR11 pKa = 11.84PTSRR15 pKa = 11.84FAKK18 pKa = 10.04KK19 pKa = 9.8PYY21 pKa = 9.81GGSKK25 pKa = 9.54SRR27 pKa = 11.84RR28 pKa = 11.84SFVKK32 pKa = 9.78RR33 pKa = 11.84RR34 pKa = 11.84TFKK37 pKa = 10.5KK38 pKa = 9.92RR39 pKa = 11.84RR40 pKa = 11.84PTMTSKK46 pKa = 11.08RR47 pKa = 11.84ILNLTATKK55 pKa = 10.5KK56 pKa = 10.2RR57 pKa = 11.84DD58 pKa = 3.32TMLTYY63 pKa = 10.89TNVSSDD69 pKa = 3.26AANGGTTYY77 pKa = 9.64RR78 pKa = 11.84TDD80 pKa = 3.11VPAVLTGGQDD90 pKa = 3.62VNSVRR95 pKa = 11.84ALLWCATARR104 pKa = 11.84DD105 pKa = 4.09NTISSANNRR114 pKa = 11.84GTAFDD119 pKa = 3.49QAARR123 pKa = 11.84TSTTPYY129 pKa = 10.13MIGLKK134 pKa = 9.77EE135 pKa = 4.24AIEE138 pKa = 4.29IQVSTGMPWQWRR150 pKa = 11.84RR151 pKa = 11.84ICFTYY156 pKa = 10.13KK157 pKa = 10.83GPIGLALGTSQNISTEE173 pKa = 4.1TSNGWVRR180 pKa = 11.84LVNDD184 pKa = 3.1ITGNPGGGQQYY195 pKa = 11.02ILFEE199 pKa = 4.4KK200 pKa = 10.4LFKK203 pKa = 10.71GQNTQDD209 pKa = 3.02WNDD212 pKa = 3.03AMTAKK217 pKa = 10.2VDD219 pKa = 3.61NTRR222 pKa = 11.84VTLKK226 pKa = 10.5YY227 pKa = 11.01DD228 pKa = 3.52KK229 pKa = 10.49TCTLASGNEE238 pKa = 3.83DD239 pKa = 3.36GFIRR243 pKa = 11.84KK244 pKa = 8.39YY245 pKa = 10.98NKK247 pKa = 7.46WHH249 pKa = 6.95PMRR252 pKa = 11.84KK253 pKa = 7.02TLVYY257 pKa = 10.82NDD259 pKa = 4.57DD260 pKa = 3.74EE261 pKa = 5.53LGGGEE266 pKa = 3.97AAQVFSTQGKK276 pKa = 10.1AGMGDD281 pKa = 4.27YY282 pKa = 10.87YY283 pKa = 11.67VLDD286 pKa = 4.14LFRR289 pKa = 11.84ARR291 pKa = 11.84TGSTSSDD298 pKa = 3.1QLSFRR303 pKa = 11.84PEE305 pKa = 3.68STLYY309 pKa = 9.33WHH311 pKa = 7.11EE312 pKa = 4.13KK313 pKa = 9.32
MM1 pKa = 7.82AYY3 pKa = 10.28ARR5 pKa = 11.84RR6 pKa = 11.84NRR8 pKa = 11.84YY9 pKa = 8.67RR10 pKa = 11.84RR11 pKa = 11.84PTSRR15 pKa = 11.84FAKK18 pKa = 10.04KK19 pKa = 9.8PYY21 pKa = 9.81GGSKK25 pKa = 9.54SRR27 pKa = 11.84RR28 pKa = 11.84SFVKK32 pKa = 9.78RR33 pKa = 11.84RR34 pKa = 11.84TFKK37 pKa = 10.5KK38 pKa = 9.92RR39 pKa = 11.84RR40 pKa = 11.84PTMTSKK46 pKa = 11.08RR47 pKa = 11.84ILNLTATKK55 pKa = 10.5KK56 pKa = 10.2RR57 pKa = 11.84DD58 pKa = 3.32TMLTYY63 pKa = 10.89TNVSSDD69 pKa = 3.26AANGGTTYY77 pKa = 9.64RR78 pKa = 11.84TDD80 pKa = 3.11VPAVLTGGQDD90 pKa = 3.62VNSVRR95 pKa = 11.84ALLWCATARR104 pKa = 11.84DD105 pKa = 4.09NTISSANNRR114 pKa = 11.84GTAFDD119 pKa = 3.49QAARR123 pKa = 11.84TSTTPYY129 pKa = 10.13MIGLKK134 pKa = 9.77EE135 pKa = 4.24AIEE138 pKa = 4.29IQVSTGMPWQWRR150 pKa = 11.84RR151 pKa = 11.84ICFTYY156 pKa = 10.13KK157 pKa = 10.83GPIGLALGTSQNISTEE173 pKa = 4.1TSNGWVRR180 pKa = 11.84LVNDD184 pKa = 3.1ITGNPGGGQQYY195 pKa = 11.02ILFEE199 pKa = 4.4KK200 pKa = 10.4LFKK203 pKa = 10.71GQNTQDD209 pKa = 3.02WNDD212 pKa = 3.03AMTAKK217 pKa = 10.2VDD219 pKa = 3.61NTRR222 pKa = 11.84VTLKK226 pKa = 10.5YY227 pKa = 11.01DD228 pKa = 3.52KK229 pKa = 10.49TCTLASGNEE238 pKa = 3.83DD239 pKa = 3.36GFIRR243 pKa = 11.84KK244 pKa = 8.39YY245 pKa = 10.98NKK247 pKa = 7.46WHH249 pKa = 6.95PMRR252 pKa = 11.84KK253 pKa = 7.02TLVYY257 pKa = 10.82NDD259 pKa = 4.57DD260 pKa = 3.74EE261 pKa = 5.53LGGGEE266 pKa = 3.97AAQVFSTQGKK276 pKa = 10.1AGMGDD281 pKa = 4.27YY282 pKa = 10.87YY283 pKa = 11.67VLDD286 pKa = 4.14LFRR289 pKa = 11.84ARR291 pKa = 11.84TGSTSSDD298 pKa = 3.1QLSFRR303 pKa = 11.84PEE305 pKa = 3.68STLYY309 pKa = 9.33WHH311 pKa = 7.11EE312 pKa = 4.13KK313 pKa = 9.32
Molecular weight: 35.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
606 |
293 |
313 |
303.0 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.086 ± 0.533 | 1.65 ± 0.5 |
6.931 ± 1.084 | 5.116 ± 1.619 |
4.455 ± 0.68 | 8.416 ± 0.152 |
1.815 ± 0.85 | 4.785 ± 0.919 |
5.941 ± 0.556 | 5.941 ± 0.325 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.815 ± 0.535 | 4.785 ± 0.698 |
3.795 ± 0.434 | 2.97 ± 0.624 |
8.251 ± 0.502 | 6.106 ± 0.667 |
7.426 ± 2.946 | 4.785 ± 0.226 |
2.805 ± 0.411 | 4.125 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
