
Beihai narna-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.35
Get precalculated fractions of proteins
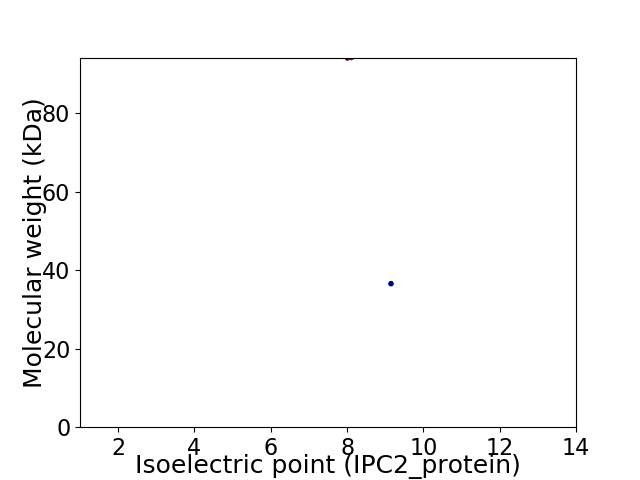
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIF2|A0A1L3KIF2_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 8 OX=1922460 PE=4 SV=1
MM1 pKa = 7.85PEE3 pKa = 3.89RR4 pKa = 11.84VSRR7 pKa = 11.84TDD9 pKa = 3.48LGPILTRR16 pKa = 11.84FGIDD20 pKa = 3.46FQSKK24 pKa = 10.06SSHH27 pKa = 6.18LLLLFSRR34 pKa = 11.84EE35 pKa = 3.75EE36 pKa = 3.58RR37 pKa = 11.84DD38 pKa = 3.45YY39 pKa = 11.86AFSAFAFLRR48 pKa = 11.84SLTEE52 pKa = 4.02CLSDD56 pKa = 3.39ADD58 pKa = 5.01RR59 pKa = 11.84VLEE62 pKa = 4.59CPDD65 pKa = 3.3VLDD68 pKa = 4.85FVPTYY73 pKa = 9.68RR74 pKa = 11.84WLMDD78 pKa = 3.06VHH80 pKa = 8.14SGEE83 pKa = 4.31EE84 pKa = 3.71QVKK87 pKa = 10.12RR88 pKa = 11.84MKK90 pKa = 10.62FLTAWPMARR99 pKa = 11.84WLRR102 pKa = 11.84NPLPPPPPRR111 pKa = 11.84VDD113 pKa = 3.14SDD115 pKa = 3.5HH116 pKa = 6.71FRR118 pKa = 11.84LPFAGRR124 pKa = 11.84THH126 pKa = 6.27RR127 pKa = 11.84HH128 pKa = 5.18LRR130 pKa = 11.84NLLASRR136 pKa = 11.84TTSVRR141 pKa = 11.84SCTVFVALLQGVKK154 pKa = 10.24RR155 pKa = 11.84GCAEE159 pKa = 3.8VNEE162 pKa = 4.43EE163 pKa = 4.42FISAALAKK171 pKa = 9.76HH172 pKa = 4.67QQALRR177 pKa = 11.84TPIPALDD184 pKa = 4.32PDD186 pKa = 3.59TRR188 pKa = 11.84RR189 pKa = 11.84NVRR192 pKa = 11.84AKK194 pKa = 10.6FSALWRR200 pKa = 11.84RR201 pKa = 11.84EE202 pKa = 4.22TPSRR206 pKa = 11.84QYY208 pKa = 11.59GEE210 pKa = 3.77WKK212 pKa = 9.92KK213 pKa = 11.0VYY215 pKa = 10.09NRR217 pKa = 11.84EE218 pKa = 4.03GKK220 pKa = 10.08CRR222 pKa = 11.84IARR225 pKa = 11.84RR226 pKa = 11.84EE227 pKa = 4.1HH228 pKa = 5.61NCSIKK233 pKa = 10.82ASFGYY238 pKa = 10.22SRR240 pKa = 11.84GAGGRR245 pKa = 11.84VGALARR251 pKa = 11.84LAKK254 pKa = 10.33DD255 pKa = 3.41YY256 pKa = 7.62WQCGHH261 pKa = 6.56NEE263 pKa = 4.45LLSMDD268 pKa = 4.41YY269 pKa = 10.66DD270 pKa = 3.88PSSGRR275 pKa = 11.84VVTQYY280 pKa = 11.09GIPSLGYY287 pKa = 9.62AALGKK292 pKa = 9.74LALMEE297 pKa = 4.18VSDD300 pKa = 5.06PQGQCHH306 pKa = 5.7ATDD309 pKa = 3.38ALLYY313 pKa = 7.42RR314 pKa = 11.84TPEE317 pKa = 4.16LLRR320 pKa = 11.84GDD322 pKa = 4.62GLCSADD328 pKa = 3.47VKK330 pKa = 10.99PILEE334 pKa = 4.16PLKK337 pKa = 10.84CRR339 pKa = 11.84LITAGSPVSYY349 pKa = 9.45AYY351 pKa = 11.11SMSAQKK357 pKa = 11.11SMWRR361 pKa = 11.84HH362 pKa = 4.56LHH364 pKa = 7.35DD365 pKa = 4.71NFPQFKK371 pKa = 10.51LIGSPLVEE379 pKa = 6.09DD380 pKa = 3.75DD381 pKa = 4.47LGWVRR386 pKa = 11.84GQTSSLDD393 pKa = 3.52LPFDD397 pKa = 3.26QWVSGDD403 pKa = 3.54YY404 pKa = 10.74SAATDD409 pKa = 3.93GLGSEE414 pKa = 4.64INRR417 pKa = 11.84LCIDD421 pKa = 4.43AYY423 pKa = 10.96CDD425 pKa = 3.38AVQATPTEE433 pKa = 4.19RR434 pKa = 11.84KK435 pKa = 8.14IWRR438 pKa = 11.84AVSGNHH444 pKa = 6.28SIHH447 pKa = 5.26YY448 pKa = 8.71TGEE451 pKa = 4.04KK452 pKa = 9.85VADD455 pKa = 3.55AVTASVGSSDD465 pKa = 3.88PFLMEE470 pKa = 4.38NGQLMGSVLSFPLLCAINLVAYY492 pKa = 7.74WAALEE497 pKa = 4.25EE498 pKa = 4.34FTGRR502 pKa = 11.84RR503 pKa = 11.84FQPNDD508 pKa = 3.27LPVLVNGDD516 pKa = 4.72DD517 pKa = 3.61ICFLANDD524 pKa = 4.03AFYY527 pKa = 10.87KK528 pKa = 10.26VWQDD532 pKa = 3.12WISRR536 pKa = 11.84VGFTLSLGKK545 pKa = 10.39NYY547 pKa = 9.84ISPDD551 pKa = 3.15FVTINSEE558 pKa = 3.9SFMYY562 pKa = 9.66SQRR565 pKa = 11.84RR566 pKa = 11.84CTFRR570 pKa = 11.84RR571 pKa = 11.84IGFLNTGLLIPSASIRR587 pKa = 11.84PEE589 pKa = 3.64NRR591 pKa = 11.84RR592 pKa = 11.84MPLTGKK598 pKa = 9.36LQRR601 pKa = 11.84LLSEE605 pKa = 4.59CNNPEE610 pKa = 3.87RR611 pKa = 11.84TWRR614 pKa = 11.84RR615 pKa = 11.84FIHH618 pKa = 5.56YY619 pKa = 9.67NKK621 pKa = 10.36AALRR625 pKa = 11.84DD626 pKa = 3.59LTLNGNVNVFGANALGSLGVTPPPGLEE653 pKa = 3.95PTYY656 pKa = 10.0TAYY659 pKa = 9.6QKK661 pKa = 11.4KK662 pKa = 7.76MAKK665 pKa = 9.63FLYY668 pKa = 9.87DD669 pKa = 3.09RR670 pKa = 11.84HH671 pKa = 5.88LATWADD677 pKa = 3.51NEE679 pKa = 4.18EE680 pKa = 4.24AKK682 pKa = 10.94VGSVWKK688 pKa = 10.18NVPPKK693 pKa = 8.98EE694 pKa = 4.09HH695 pKa = 7.25KK696 pKa = 10.95LPGSYY701 pKa = 10.57LLEE704 pKa = 4.8GCDD707 pKa = 4.92DD708 pKa = 3.69ATILAKK714 pKa = 10.26KK715 pKa = 9.4KK716 pKa = 9.76HH717 pKa = 6.27LGTEE721 pKa = 3.53RR722 pKa = 11.84CYY724 pKa = 10.5PVGLVHH730 pKa = 6.64EE731 pKa = 5.06SEE733 pKa = 3.81WSEE736 pKa = 4.14EE737 pKa = 4.14NEE739 pKa = 4.32LPLPAALPALNYY751 pKa = 10.23QLFGGPLTYY760 pKa = 10.59FGDD763 pKa = 3.99VSWSVKK769 pKa = 10.2YY770 pKa = 10.65LSKK773 pKa = 11.13ADD775 pKa = 3.57LRR777 pKa = 11.84SFRR780 pKa = 11.84SCLRR784 pKa = 11.84RR785 pKa = 11.84AEE787 pKa = 4.08RR788 pKa = 11.84DD789 pKa = 3.28PRR791 pKa = 11.84EE792 pKa = 3.57WKK794 pKa = 10.15YY795 pKa = 11.28RR796 pKa = 11.84LGRR799 pKa = 11.84MSTDD803 pKa = 3.06EE804 pKa = 4.11TPATNKK810 pKa = 9.92LRR812 pKa = 11.84VEE814 pKa = 4.66MPTEE818 pKa = 4.11PPPTGVEE825 pKa = 4.44GPADD829 pKa = 3.71SVRR832 pKa = 11.84RR833 pKa = 11.84WW834 pKa = 3.0
MM1 pKa = 7.85PEE3 pKa = 3.89RR4 pKa = 11.84VSRR7 pKa = 11.84TDD9 pKa = 3.48LGPILTRR16 pKa = 11.84FGIDD20 pKa = 3.46FQSKK24 pKa = 10.06SSHH27 pKa = 6.18LLLLFSRR34 pKa = 11.84EE35 pKa = 3.75EE36 pKa = 3.58RR37 pKa = 11.84DD38 pKa = 3.45YY39 pKa = 11.86AFSAFAFLRR48 pKa = 11.84SLTEE52 pKa = 4.02CLSDD56 pKa = 3.39ADD58 pKa = 5.01RR59 pKa = 11.84VLEE62 pKa = 4.59CPDD65 pKa = 3.3VLDD68 pKa = 4.85FVPTYY73 pKa = 9.68RR74 pKa = 11.84WLMDD78 pKa = 3.06VHH80 pKa = 8.14SGEE83 pKa = 4.31EE84 pKa = 3.71QVKK87 pKa = 10.12RR88 pKa = 11.84MKK90 pKa = 10.62FLTAWPMARR99 pKa = 11.84WLRR102 pKa = 11.84NPLPPPPPRR111 pKa = 11.84VDD113 pKa = 3.14SDD115 pKa = 3.5HH116 pKa = 6.71FRR118 pKa = 11.84LPFAGRR124 pKa = 11.84THH126 pKa = 6.27RR127 pKa = 11.84HH128 pKa = 5.18LRR130 pKa = 11.84NLLASRR136 pKa = 11.84TTSVRR141 pKa = 11.84SCTVFVALLQGVKK154 pKa = 10.24RR155 pKa = 11.84GCAEE159 pKa = 3.8VNEE162 pKa = 4.43EE163 pKa = 4.42FISAALAKK171 pKa = 9.76HH172 pKa = 4.67QQALRR177 pKa = 11.84TPIPALDD184 pKa = 4.32PDD186 pKa = 3.59TRR188 pKa = 11.84RR189 pKa = 11.84NVRR192 pKa = 11.84AKK194 pKa = 10.6FSALWRR200 pKa = 11.84RR201 pKa = 11.84EE202 pKa = 4.22TPSRR206 pKa = 11.84QYY208 pKa = 11.59GEE210 pKa = 3.77WKK212 pKa = 9.92KK213 pKa = 11.0VYY215 pKa = 10.09NRR217 pKa = 11.84EE218 pKa = 4.03GKK220 pKa = 10.08CRR222 pKa = 11.84IARR225 pKa = 11.84RR226 pKa = 11.84EE227 pKa = 4.1HH228 pKa = 5.61NCSIKK233 pKa = 10.82ASFGYY238 pKa = 10.22SRR240 pKa = 11.84GAGGRR245 pKa = 11.84VGALARR251 pKa = 11.84LAKK254 pKa = 10.33DD255 pKa = 3.41YY256 pKa = 7.62WQCGHH261 pKa = 6.56NEE263 pKa = 4.45LLSMDD268 pKa = 4.41YY269 pKa = 10.66DD270 pKa = 3.88PSSGRR275 pKa = 11.84VVTQYY280 pKa = 11.09GIPSLGYY287 pKa = 9.62AALGKK292 pKa = 9.74LALMEE297 pKa = 4.18VSDD300 pKa = 5.06PQGQCHH306 pKa = 5.7ATDD309 pKa = 3.38ALLYY313 pKa = 7.42RR314 pKa = 11.84TPEE317 pKa = 4.16LLRR320 pKa = 11.84GDD322 pKa = 4.62GLCSADD328 pKa = 3.47VKK330 pKa = 10.99PILEE334 pKa = 4.16PLKK337 pKa = 10.84CRR339 pKa = 11.84LITAGSPVSYY349 pKa = 9.45AYY351 pKa = 11.11SMSAQKK357 pKa = 11.11SMWRR361 pKa = 11.84HH362 pKa = 4.56LHH364 pKa = 7.35DD365 pKa = 4.71NFPQFKK371 pKa = 10.51LIGSPLVEE379 pKa = 6.09DD380 pKa = 3.75DD381 pKa = 4.47LGWVRR386 pKa = 11.84GQTSSLDD393 pKa = 3.52LPFDD397 pKa = 3.26QWVSGDD403 pKa = 3.54YY404 pKa = 10.74SAATDD409 pKa = 3.93GLGSEE414 pKa = 4.64INRR417 pKa = 11.84LCIDD421 pKa = 4.43AYY423 pKa = 10.96CDD425 pKa = 3.38AVQATPTEE433 pKa = 4.19RR434 pKa = 11.84KK435 pKa = 8.14IWRR438 pKa = 11.84AVSGNHH444 pKa = 6.28SIHH447 pKa = 5.26YY448 pKa = 8.71TGEE451 pKa = 4.04KK452 pKa = 9.85VADD455 pKa = 3.55AVTASVGSSDD465 pKa = 3.88PFLMEE470 pKa = 4.38NGQLMGSVLSFPLLCAINLVAYY492 pKa = 7.74WAALEE497 pKa = 4.25EE498 pKa = 4.34FTGRR502 pKa = 11.84RR503 pKa = 11.84FQPNDD508 pKa = 3.27LPVLVNGDD516 pKa = 4.72DD517 pKa = 3.61ICFLANDD524 pKa = 4.03AFYY527 pKa = 10.87KK528 pKa = 10.26VWQDD532 pKa = 3.12WISRR536 pKa = 11.84VGFTLSLGKK545 pKa = 10.39NYY547 pKa = 9.84ISPDD551 pKa = 3.15FVTINSEE558 pKa = 3.9SFMYY562 pKa = 9.66SQRR565 pKa = 11.84RR566 pKa = 11.84CTFRR570 pKa = 11.84RR571 pKa = 11.84IGFLNTGLLIPSASIRR587 pKa = 11.84PEE589 pKa = 3.64NRR591 pKa = 11.84RR592 pKa = 11.84MPLTGKK598 pKa = 9.36LQRR601 pKa = 11.84LLSEE605 pKa = 4.59CNNPEE610 pKa = 3.87RR611 pKa = 11.84TWRR614 pKa = 11.84RR615 pKa = 11.84FIHH618 pKa = 5.56YY619 pKa = 9.67NKK621 pKa = 10.36AALRR625 pKa = 11.84DD626 pKa = 3.59LTLNGNVNVFGANALGSLGVTPPPGLEE653 pKa = 3.95PTYY656 pKa = 10.0TAYY659 pKa = 9.6QKK661 pKa = 11.4KK662 pKa = 7.76MAKK665 pKa = 9.63FLYY668 pKa = 9.87DD669 pKa = 3.09RR670 pKa = 11.84HH671 pKa = 5.88LATWADD677 pKa = 3.51NEE679 pKa = 4.18EE680 pKa = 4.24AKK682 pKa = 10.94VGSVWKK688 pKa = 10.18NVPPKK693 pKa = 8.98EE694 pKa = 4.09HH695 pKa = 7.25KK696 pKa = 10.95LPGSYY701 pKa = 10.57LLEE704 pKa = 4.8GCDD707 pKa = 4.92DD708 pKa = 3.69ATILAKK714 pKa = 10.26KK715 pKa = 9.4KK716 pKa = 9.76HH717 pKa = 6.27LGTEE721 pKa = 3.53RR722 pKa = 11.84CYY724 pKa = 10.5PVGLVHH730 pKa = 6.64EE731 pKa = 5.06SEE733 pKa = 3.81WSEE736 pKa = 4.14EE737 pKa = 4.14NEE739 pKa = 4.32LPLPAALPALNYY751 pKa = 10.23QLFGGPLTYY760 pKa = 10.59FGDD763 pKa = 3.99VSWSVKK769 pKa = 10.2YY770 pKa = 10.65LSKK773 pKa = 11.13ADD775 pKa = 3.57LRR777 pKa = 11.84SFRR780 pKa = 11.84SCLRR784 pKa = 11.84RR785 pKa = 11.84AEE787 pKa = 4.08RR788 pKa = 11.84DD789 pKa = 3.28PRR791 pKa = 11.84EE792 pKa = 3.57WKK794 pKa = 10.15YY795 pKa = 11.28RR796 pKa = 11.84LGRR799 pKa = 11.84MSTDD803 pKa = 3.06EE804 pKa = 4.11TPATNKK810 pKa = 9.92LRR812 pKa = 11.84VEE814 pKa = 4.66MPTEE818 pKa = 4.11PPPTGVEE825 pKa = 4.44GPADD829 pKa = 3.71SVRR832 pKa = 11.84RR833 pKa = 11.84WW834 pKa = 3.0
Molecular weight: 94.06 kDa
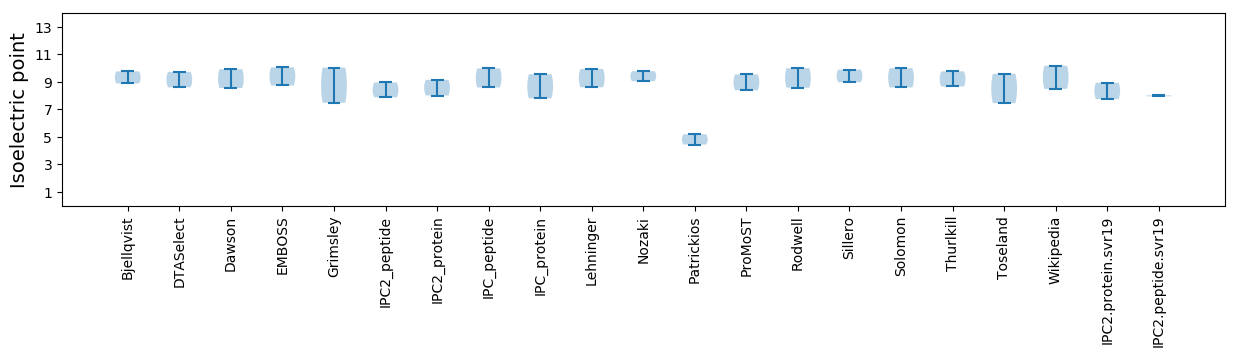
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIF2|A0A1L3KIF2_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 8 OX=1922460 PE=4 SV=1
MM1 pKa = 7.62AANNKK6 pKa = 7.67KK7 pKa = 7.14TTVTTTTTQSNGASIPRR24 pKa = 11.84RR25 pKa = 11.84WGRR28 pKa = 11.84AARR31 pKa = 11.84RR32 pKa = 11.84PLEE35 pKa = 4.4GIKK38 pKa = 10.21QGVGQVTIEE47 pKa = 4.08AFPRR51 pKa = 11.84AARR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84TGNPMRR66 pKa = 11.84GYY68 pKa = 10.78FNALSPHH75 pKa = 6.2HH76 pKa = 6.7LALPRR81 pKa = 11.84ATGPYY86 pKa = 8.14MVCRR90 pKa = 11.84TTTRR94 pKa = 11.84FSSTGVVHH102 pKa = 7.13LLGPMTAIPVAQTEE116 pKa = 4.74SPWAAVVGVTHH127 pKa = 7.2PGGGHH132 pKa = 4.94VNPMNGINNVIPIIDD147 pKa = 3.84AGLNVSGFNDD157 pKa = 3.52SRR159 pKa = 11.84VTPAAFTVQVMNPGALQTTSGIVYY183 pKa = 9.58QGRR186 pKa = 11.84VNNVPALMDD195 pKa = 5.2DD196 pKa = 3.49PRR198 pKa = 11.84TWAAYY203 pKa = 10.09ADD205 pKa = 4.08SLVSFCSPRR214 pKa = 11.84MCSAGKK220 pKa = 9.56LALRR224 pKa = 11.84GTQTSLVPFNMSKK237 pKa = 10.46LAEE240 pKa = 4.07FSEE243 pKa = 4.84RR244 pKa = 11.84SLVVPGATTWNPGTLGLQFQGFAPMFVYY272 pKa = 10.66NPDD275 pKa = 4.2GIEE278 pKa = 3.91LEE280 pKa = 4.37YY281 pKa = 11.05VVTIEE286 pKa = 3.44WRR288 pKa = 11.84VRR290 pKa = 11.84FDD292 pKa = 3.66PSNPAHH298 pKa = 6.8AGSEE302 pKa = 4.12YY303 pKa = 10.45HH304 pKa = 6.75PPSSDD309 pKa = 3.68SLWSRR314 pKa = 11.84CVEE317 pKa = 4.02AASSVGNGVEE327 pKa = 4.82DD328 pKa = 3.56IADD331 pKa = 3.42VVAAVGAAAALLL343 pKa = 4.02
MM1 pKa = 7.62AANNKK6 pKa = 7.67KK7 pKa = 7.14TTVTTTTTQSNGASIPRR24 pKa = 11.84RR25 pKa = 11.84WGRR28 pKa = 11.84AARR31 pKa = 11.84RR32 pKa = 11.84PLEE35 pKa = 4.4GIKK38 pKa = 10.21QGVGQVTIEE47 pKa = 4.08AFPRR51 pKa = 11.84AARR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84TGNPMRR66 pKa = 11.84GYY68 pKa = 10.78FNALSPHH75 pKa = 6.2HH76 pKa = 6.7LALPRR81 pKa = 11.84ATGPYY86 pKa = 8.14MVCRR90 pKa = 11.84TTTRR94 pKa = 11.84FSSTGVVHH102 pKa = 7.13LLGPMTAIPVAQTEE116 pKa = 4.74SPWAAVVGVTHH127 pKa = 7.2PGGGHH132 pKa = 4.94VNPMNGINNVIPIIDD147 pKa = 3.84AGLNVSGFNDD157 pKa = 3.52SRR159 pKa = 11.84VTPAAFTVQVMNPGALQTTSGIVYY183 pKa = 9.58QGRR186 pKa = 11.84VNNVPALMDD195 pKa = 5.2DD196 pKa = 3.49PRR198 pKa = 11.84TWAAYY203 pKa = 10.09ADD205 pKa = 4.08SLVSFCSPRR214 pKa = 11.84MCSAGKK220 pKa = 9.56LALRR224 pKa = 11.84GTQTSLVPFNMSKK237 pKa = 10.46LAEE240 pKa = 4.07FSEE243 pKa = 4.84RR244 pKa = 11.84SLVVPGATTWNPGTLGLQFQGFAPMFVYY272 pKa = 10.66NPDD275 pKa = 4.2GIEE278 pKa = 3.91LEE280 pKa = 4.37YY281 pKa = 11.05VVTIEE286 pKa = 3.44WRR288 pKa = 11.84VRR290 pKa = 11.84FDD292 pKa = 3.66PSNPAHH298 pKa = 6.8AGSEE302 pKa = 4.12YY303 pKa = 10.45HH304 pKa = 6.75PPSSDD309 pKa = 3.68SLWSRR314 pKa = 11.84CVEE317 pKa = 4.02AASSVGNGVEE327 pKa = 4.82DD328 pKa = 3.56IADD331 pKa = 3.42VVAAVGAAAALLL343 pKa = 4.02
Molecular weight: 36.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1177 |
343 |
834 |
588.5 |
65.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.921 ± 1.276 | 1.954 ± 0.41 |
4.758 ± 0.96 | 4.843 ± 0.852 |
3.908 ± 0.213 | 7.392 ± 0.858 |
2.124 ± 0.043 | 3.059 ± 0.229 |
3.483 ± 1.055 | 9.686 ± 1.856 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.412 | 4.248 ± 0.673 |
7.052 ± 0.579 | 2.719 ± 0.102 |
8.071 ± 0.408 | 7.562 ± 0.142 |
5.947 ± 1.154 | 6.882 ± 1.427 |
2.209 ± 0.239 | 3.059 ± 0.53 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
