
Capybara microvirus Cap1_SP_148
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
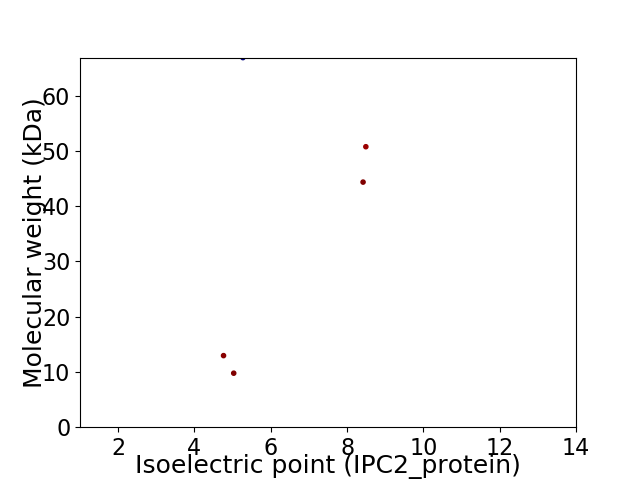
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
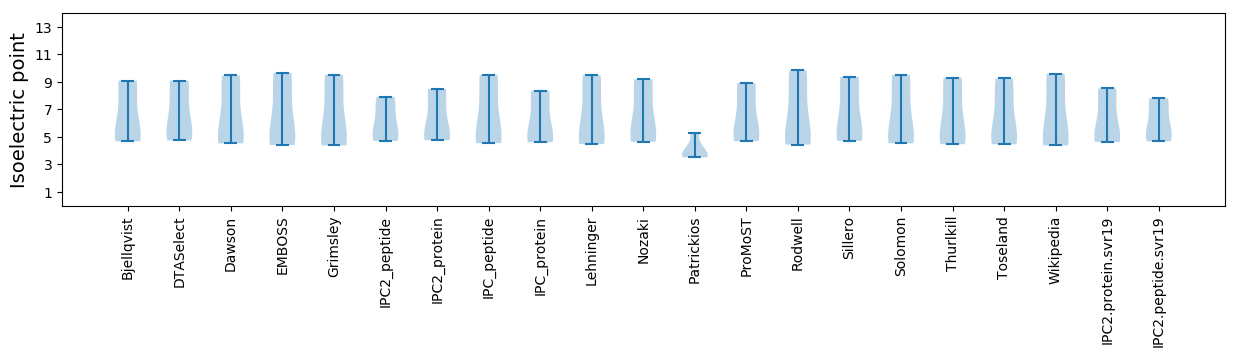
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7Y6|A0A4P8W7Y6_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_148 OX=2585392 PE=4 SV=1
MM1 pKa = 7.26CLKK4 pKa = 10.35YY5 pKa = 10.43KK6 pKa = 10.56YY7 pKa = 8.05LTPVPPLPDD16 pKa = 2.82ATFYY20 pKa = 11.65EE21 pKa = 5.09EE22 pKa = 4.06VHH24 pKa = 6.12TPNGYY29 pKa = 9.42KK30 pKa = 10.24VSEE33 pKa = 4.35EE34 pKa = 3.81PLYY37 pKa = 10.47PPVSAFSIEE46 pKa = 4.01NLLKK50 pKa = 10.73SGIQIYY56 pKa = 9.09PSPYY60 pKa = 8.69LRR62 pKa = 11.84PMSRR66 pKa = 11.84FNVLSSAEE74 pKa = 4.05RR75 pKa = 11.84FLNTPEE81 pKa = 4.61LVEE84 pKa = 5.57ASQKK88 pKa = 8.39WSEE91 pKa = 4.03MTEE94 pKa = 4.06EE95 pKa = 4.71FLSQAQKK102 pKa = 10.6IDD104 pKa = 3.56TSNDD108 pKa = 3.23LPDD111 pKa = 4.84DD112 pKa = 3.88KK113 pKa = 11.7
MM1 pKa = 7.26CLKK4 pKa = 10.35YY5 pKa = 10.43KK6 pKa = 10.56YY7 pKa = 8.05LTPVPPLPDD16 pKa = 2.82ATFYY20 pKa = 11.65EE21 pKa = 5.09EE22 pKa = 4.06VHH24 pKa = 6.12TPNGYY29 pKa = 9.42KK30 pKa = 10.24VSEE33 pKa = 4.35EE34 pKa = 3.81PLYY37 pKa = 10.47PPVSAFSIEE46 pKa = 4.01NLLKK50 pKa = 10.73SGIQIYY56 pKa = 9.09PSPYY60 pKa = 8.69LRR62 pKa = 11.84PMSRR66 pKa = 11.84FNVLSSAEE74 pKa = 4.05RR75 pKa = 11.84FLNTPEE81 pKa = 4.61LVEE84 pKa = 5.57ASQKK88 pKa = 8.39WSEE91 pKa = 4.03MTEE94 pKa = 4.06EE95 pKa = 4.71FLSQAQKK102 pKa = 10.6IDD104 pKa = 3.56TSNDD108 pKa = 3.23LPDD111 pKa = 4.84DD112 pKa = 3.88KK113 pKa = 11.7
Molecular weight: 12.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5G6|A0A4P8W5G6_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_148 OX=2585392 PE=4 SV=1
MM1 pKa = 7.39SLLGGILGGASSLLGGAINAFTAYY25 pKa = 10.43DD26 pKa = 3.68LQNQQFAEE34 pKa = 4.18QAKK37 pKa = 9.44QRR39 pKa = 11.84DD40 pKa = 3.57WQEE43 pKa = 3.93RR44 pKa = 11.84MMDD47 pKa = 4.0KK48 pKa = 10.79EE49 pKa = 3.92NAYY52 pKa = 8.33NTDD55 pKa = 2.9MWNKK59 pKa = 8.41TSEE62 pKa = 4.13FNEE65 pKa = 3.67RR66 pKa = 11.84MMRR69 pKa = 11.84AQMEE73 pKa = 4.42NASRR77 pKa = 11.84MQTEE81 pKa = 3.98QNVWNSEE88 pKa = 3.66QAQYY92 pKa = 10.97QRR94 pKa = 11.84YY95 pKa = 8.43KK96 pKa = 10.5ATGLNPLGFGGNSSTSPAIQAGSASMSPQSSAAPPMPAQYY136 pKa = 11.65ANMINPRR143 pKa = 11.84WGDD146 pKa = 3.41MLTNIAQGAEE156 pKa = 3.84LGKK159 pKa = 10.84VFGEE163 pKa = 4.08TRR165 pKa = 11.84KK166 pKa = 10.49LKK168 pKa = 10.89EE169 pKa = 3.6EE170 pKa = 4.15TKK172 pKa = 10.65NIEE175 pKa = 3.57VDD177 pKa = 3.34RR178 pKa = 11.84LKK180 pKa = 11.13KK181 pKa = 10.54LVEE184 pKa = 4.09TQLNKK189 pKa = 10.74AQIAKK194 pKa = 9.82LAEE197 pKa = 4.15EE198 pKa = 4.43VVEE201 pKa = 4.17TRR203 pKa = 11.84QRR205 pKa = 11.84ISQINLDD212 pKa = 3.94NEE214 pKa = 4.01AKK216 pKa = 10.51EE217 pKa = 4.43IINKK221 pKa = 9.74YY222 pKa = 9.61LDD224 pKa = 3.79RR225 pKa = 11.84NSALQLRR232 pKa = 11.84HH233 pKa = 6.52LEE235 pKa = 4.16LEE237 pKa = 3.98NLQTQKK243 pKa = 9.05QTDD246 pKa = 3.97KK247 pKa = 10.81IKK249 pKa = 11.22SEE251 pKa = 3.55ISKK254 pKa = 10.36IKK256 pKa = 9.86TDD258 pKa = 3.14ISLSEE263 pKa = 4.09SEE265 pKa = 4.24RR266 pKa = 11.84KK267 pKa = 9.65KK268 pKa = 10.73VEE270 pKa = 3.94QFVSQQSKK278 pKa = 10.45LFPQEE283 pKa = 3.92LQEE286 pKa = 4.63KK287 pKa = 9.06FLKK290 pKa = 10.86NEE292 pKa = 4.04LSSKK296 pKa = 10.07QSQEE300 pKa = 3.78FSNKK304 pKa = 9.54LKK306 pKa = 10.13TFDD309 pKa = 4.91LEE311 pKa = 4.24YY312 pKa = 10.97EE313 pKa = 4.11KK314 pKa = 11.14LKK316 pKa = 11.38NEE318 pKa = 4.57LKK320 pKa = 10.97LSDD323 pKa = 3.96AQLLDD328 pKa = 3.87LLRR331 pKa = 11.84VEE333 pKa = 4.66QNLKK337 pKa = 10.3IPFFGNFKK345 pKa = 10.5FYY347 pKa = 10.84TPAKK351 pKa = 9.56LKK353 pKa = 10.38EE354 pKa = 4.05NKK356 pKa = 9.52LMKK359 pKa = 10.12NQQSRR364 pKa = 11.84YY365 pKa = 9.48NIRR368 pKa = 11.84SDD370 pKa = 3.02PRR372 pKa = 11.84RR373 pKa = 11.84RR374 pKa = 11.84DD375 pKa = 3.45FMDD378 pKa = 4.4FPSYY382 pKa = 11.28VNPP385 pKa = 4.2
MM1 pKa = 7.39SLLGGILGGASSLLGGAINAFTAYY25 pKa = 10.43DD26 pKa = 3.68LQNQQFAEE34 pKa = 4.18QAKK37 pKa = 9.44QRR39 pKa = 11.84DD40 pKa = 3.57WQEE43 pKa = 3.93RR44 pKa = 11.84MMDD47 pKa = 4.0KK48 pKa = 10.79EE49 pKa = 3.92NAYY52 pKa = 8.33NTDD55 pKa = 2.9MWNKK59 pKa = 8.41TSEE62 pKa = 4.13FNEE65 pKa = 3.67RR66 pKa = 11.84MMRR69 pKa = 11.84AQMEE73 pKa = 4.42NASRR77 pKa = 11.84MQTEE81 pKa = 3.98QNVWNSEE88 pKa = 3.66QAQYY92 pKa = 10.97QRR94 pKa = 11.84YY95 pKa = 8.43KK96 pKa = 10.5ATGLNPLGFGGNSSTSPAIQAGSASMSPQSSAAPPMPAQYY136 pKa = 11.65ANMINPRR143 pKa = 11.84WGDD146 pKa = 3.41MLTNIAQGAEE156 pKa = 3.84LGKK159 pKa = 10.84VFGEE163 pKa = 4.08TRR165 pKa = 11.84KK166 pKa = 10.49LKK168 pKa = 10.89EE169 pKa = 3.6EE170 pKa = 4.15TKK172 pKa = 10.65NIEE175 pKa = 3.57VDD177 pKa = 3.34RR178 pKa = 11.84LKK180 pKa = 11.13KK181 pKa = 10.54LVEE184 pKa = 4.09TQLNKK189 pKa = 10.74AQIAKK194 pKa = 9.82LAEE197 pKa = 4.15EE198 pKa = 4.43VVEE201 pKa = 4.17TRR203 pKa = 11.84QRR205 pKa = 11.84ISQINLDD212 pKa = 3.94NEE214 pKa = 4.01AKK216 pKa = 10.51EE217 pKa = 4.43IINKK221 pKa = 9.74YY222 pKa = 9.61LDD224 pKa = 3.79RR225 pKa = 11.84NSALQLRR232 pKa = 11.84HH233 pKa = 6.52LEE235 pKa = 4.16LEE237 pKa = 3.98NLQTQKK243 pKa = 9.05QTDD246 pKa = 3.97KK247 pKa = 10.81IKK249 pKa = 11.22SEE251 pKa = 3.55ISKK254 pKa = 10.36IKK256 pKa = 9.86TDD258 pKa = 3.14ISLSEE263 pKa = 4.09SEE265 pKa = 4.24RR266 pKa = 11.84KK267 pKa = 9.65KK268 pKa = 10.73VEE270 pKa = 3.94QFVSQQSKK278 pKa = 10.45LFPQEE283 pKa = 3.92LQEE286 pKa = 4.63KK287 pKa = 9.06FLKK290 pKa = 10.86NEE292 pKa = 4.04LSSKK296 pKa = 10.07QSQEE300 pKa = 3.78FSNKK304 pKa = 9.54LKK306 pKa = 10.13TFDD309 pKa = 4.91LEE311 pKa = 4.24YY312 pKa = 10.97EE313 pKa = 4.11KK314 pKa = 11.14LKK316 pKa = 11.38NEE318 pKa = 4.57LKK320 pKa = 10.97LSDD323 pKa = 3.96AQLLDD328 pKa = 3.87LLRR331 pKa = 11.84VEE333 pKa = 4.66QNLKK337 pKa = 10.3IPFFGNFKK345 pKa = 10.5FYY347 pKa = 10.84TPAKK351 pKa = 9.56LKK353 pKa = 10.38EE354 pKa = 4.05NKK356 pKa = 9.52LMKK359 pKa = 10.12NQQSRR364 pKa = 11.84YY365 pKa = 9.48NIRR368 pKa = 11.84SDD370 pKa = 3.02PRR372 pKa = 11.84RR373 pKa = 11.84RR374 pKa = 11.84DD375 pKa = 3.45FMDD378 pKa = 4.4FPSYY382 pKa = 11.28VNPP385 pKa = 4.2
Molecular weight: 44.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1588 |
86 |
582 |
317.6 |
36.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.975 ± 0.699 | 1.071 ± 0.385 |
6.108 ± 0.744 | 5.668 ± 1.411 |
5.982 ± 0.795 | 4.156 ± 0.79 |
1.889 ± 0.684 | 4.03 ± 0.385 |
6.423 ± 1.369 | 9.824 ± 1.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.204 ± 0.529 | 6.234 ± 0.693 |
4.534 ± 0.962 | 5.227 ± 1.23 |
5.227 ± 0.825 | 7.997 ± 1.187 |
5.856 ± 0.721 | 5.353 ± 1.07 |
1.259 ± 0.206 | 5.982 ± 1.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
