
Deinococcus proteolyticus (strain ATCC 35074 / DSM 20540 / JCM 6276 / NBRC 101906 / NCIMB 13154 / VKM Ac-1939 / CCM 2703 / MRP)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus; Deinococcus proteolyticus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
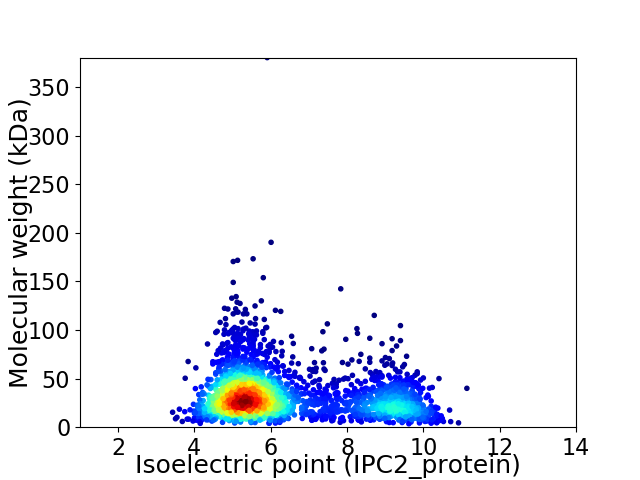
Virtual 2D-PAGE plot for 2652 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|F0RQ13|F0RQ13_DEIPM ATP phosphoribosyltransferase OS=Deinococcus proteolyticus (strain ATCC 35074 / DSM 20540 / JCM 6276 / NBRC 101906 / NCIMB 13154 / VKM Ac-1939 / CCM 2703 / MRP) OX=693977 GN=hisG PE=3 SV=1
MM1 pKa = 7.53PSKK4 pKa = 10.76SVVEE8 pKa = 4.16ALSDD12 pKa = 4.54PIYY15 pKa = 10.35SACSPDD21 pKa = 4.08LLGMFATDD29 pKa = 3.77MPSKK33 pKa = 10.65SVVEE37 pKa = 4.16ALSDD41 pKa = 4.54PIYY44 pKa = 10.35SACSPDD50 pKa = 3.9LLGMFDD56 pKa = 3.43
MM1 pKa = 7.53PSKK4 pKa = 10.76SVVEE8 pKa = 4.16ALSDD12 pKa = 4.54PIYY15 pKa = 10.35SACSPDD21 pKa = 4.08LLGMFATDD29 pKa = 3.77MPSKK33 pKa = 10.65SVVEE37 pKa = 4.16ALSDD41 pKa = 4.54PIYY44 pKa = 10.35SACSPDD50 pKa = 3.9LLGMFDD56 pKa = 3.43
Molecular weight: 5.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0RKY0|F0RKY0_DEIPM FAD dependent oxidoreductase OS=Deinococcus proteolyticus (strain ATCC 35074 / DSM 20540 / JCM 6276 / NBRC 101906 / NCIMB 13154 / VKM Ac-1939 / CCM 2703 / MRP) OX=693977 GN=Deipr_0591 PE=3 SV=1
MM1 pKa = 7.5PRR3 pKa = 11.84WKK5 pKa = 10.06SWPWPRR11 pKa = 11.84GSRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84AAAQRR21 pKa = 11.84GVQAARR27 pKa = 11.84QARR30 pKa = 11.84RR31 pKa = 11.84SVPRR35 pKa = 11.84QRR37 pKa = 11.84TVVLPGQPPEE47 pKa = 3.94RR48 pKa = 11.84ASCRR52 pKa = 11.84TCRR55 pKa = 11.84AGRR58 pKa = 11.84ASAQALHH65 pKa = 6.94LSQRR69 pKa = 11.84HH70 pKa = 4.58PQQLRR75 pKa = 11.84LDD77 pKa = 3.98QQAPPQPHH85 pKa = 5.8QLLSTRR91 pKa = 11.84LLSAQLPLLQRR102 pKa = 11.84QFRR105 pKa = 11.84IRR107 pKa = 11.84PPLPQWRR114 pKa = 11.84RR115 pKa = 11.84LPPPLHH121 pKa = 6.19RR122 pKa = 11.84QPLHH126 pKa = 5.75RR127 pKa = 11.84QPLHH131 pKa = 6.25RR132 pKa = 11.84RR133 pKa = 11.84PPRR136 pKa = 11.84HH137 pKa = 6.26PRR139 pKa = 11.84HH140 pKa = 6.65LYY142 pKa = 9.6PKK144 pKa = 8.52YY145 pKa = 9.99LSPRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84KK152 pKa = 9.65PGRR155 pKa = 11.84QPTRR159 pKa = 11.84HH160 pKa = 5.63RR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84QPLCPGRR172 pKa = 11.84TLPQLQPRR180 pKa = 11.84SPRR183 pKa = 11.84PPRR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84LPRR191 pKa = 11.84RR192 pKa = 11.84FLLLPQPLPQAQSWPRR208 pKa = 11.84HH209 pKa = 4.43RR210 pKa = 11.84QPRR213 pKa = 11.84HH214 pKa = 4.49RR215 pKa = 11.84QPRR218 pKa = 11.84RR219 pKa = 11.84QQQRR223 pKa = 11.84RR224 pKa = 11.84HH225 pKa = 4.44QQRR228 pKa = 11.84HH229 pKa = 4.04RR230 pKa = 11.84QRR232 pKa = 11.84PSQRR236 pKa = 11.84HH237 pKa = 4.39LSQRR241 pKa = 11.84CRR243 pKa = 11.84PRR245 pKa = 11.84RR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84SRR250 pKa = 11.84WLPLPPPRR258 pKa = 11.84RR259 pKa = 11.84PSRR262 pKa = 11.84WPRR265 pKa = 11.84HH266 pKa = 5.35RR267 pKa = 11.84PQPQDD272 pKa = 3.19PPLSRR277 pKa = 11.84RR278 pKa = 11.84PARR281 pKa = 11.84PQYY284 pKa = 9.23PAQPRR289 pKa = 11.84HH290 pKa = 5.73PVPSRR295 pKa = 11.84PHH297 pKa = 5.27SQQTRR302 pKa = 11.84QPLRR306 pKa = 11.84PHH308 pKa = 6.83RR309 pKa = 11.84PRR311 pKa = 11.84RR312 pKa = 11.84PLLLPPWWPQRR323 pKa = 11.84RR324 pKa = 11.84PLNQQ328 pKa = 3.02
MM1 pKa = 7.5PRR3 pKa = 11.84WKK5 pKa = 10.06SWPWPRR11 pKa = 11.84GSRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84AAAQRR21 pKa = 11.84GVQAARR27 pKa = 11.84QARR30 pKa = 11.84RR31 pKa = 11.84SVPRR35 pKa = 11.84QRR37 pKa = 11.84TVVLPGQPPEE47 pKa = 3.94RR48 pKa = 11.84ASCRR52 pKa = 11.84TCRR55 pKa = 11.84AGRR58 pKa = 11.84ASAQALHH65 pKa = 6.94LSQRR69 pKa = 11.84HH70 pKa = 4.58PQQLRR75 pKa = 11.84LDD77 pKa = 3.98QQAPPQPHH85 pKa = 5.8QLLSTRR91 pKa = 11.84LLSAQLPLLQRR102 pKa = 11.84QFRR105 pKa = 11.84IRR107 pKa = 11.84PPLPQWRR114 pKa = 11.84RR115 pKa = 11.84LPPPLHH121 pKa = 6.19RR122 pKa = 11.84QPLHH126 pKa = 5.75RR127 pKa = 11.84QPLHH131 pKa = 6.25RR132 pKa = 11.84RR133 pKa = 11.84PPRR136 pKa = 11.84HH137 pKa = 6.26PRR139 pKa = 11.84HH140 pKa = 6.65LYY142 pKa = 9.6PKK144 pKa = 8.52YY145 pKa = 9.99LSPRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84KK152 pKa = 9.65PGRR155 pKa = 11.84QPTRR159 pKa = 11.84HH160 pKa = 5.63RR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84QPLCPGRR172 pKa = 11.84TLPQLQPRR180 pKa = 11.84SPRR183 pKa = 11.84PPRR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84LPRR191 pKa = 11.84RR192 pKa = 11.84FLLLPQPLPQAQSWPRR208 pKa = 11.84HH209 pKa = 4.43RR210 pKa = 11.84QPRR213 pKa = 11.84HH214 pKa = 4.49RR215 pKa = 11.84QPRR218 pKa = 11.84RR219 pKa = 11.84QQQRR223 pKa = 11.84RR224 pKa = 11.84HH225 pKa = 4.44QQRR228 pKa = 11.84HH229 pKa = 4.04RR230 pKa = 11.84QRR232 pKa = 11.84PSQRR236 pKa = 11.84HH237 pKa = 4.39LSQRR241 pKa = 11.84CRR243 pKa = 11.84PRR245 pKa = 11.84RR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84SRR250 pKa = 11.84WLPLPPPRR258 pKa = 11.84RR259 pKa = 11.84PSRR262 pKa = 11.84WPRR265 pKa = 11.84HH266 pKa = 5.35RR267 pKa = 11.84PQPQDD272 pKa = 3.19PPLSRR277 pKa = 11.84RR278 pKa = 11.84PARR281 pKa = 11.84PQYY284 pKa = 9.23PAQPRR289 pKa = 11.84HH290 pKa = 5.73PVPSRR295 pKa = 11.84PHH297 pKa = 5.27SQQTRR302 pKa = 11.84QPLRR306 pKa = 11.84PHH308 pKa = 6.83RR309 pKa = 11.84PRR311 pKa = 11.84RR312 pKa = 11.84PLLLPPWWPQRR323 pKa = 11.84RR324 pKa = 11.84PLNQQ328 pKa = 3.02
Molecular weight: 40.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
832983 |
30 |
3676 |
314.1 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.406 ± 0.08 | 0.604 ± 0.013 |
4.905 ± 0.036 | 6.189 ± 0.055 |
2.987 ± 0.026 | 9.154 ± 0.046 |
2.087 ± 0.026 | 3.403 ± 0.038 |
2.457 ± 0.041 | 11.871 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.018 ± 0.025 | 2.325 ± 0.03 |
5.842 ± 0.04 | 4.619 ± 0.036 |
7.152 ± 0.052 | 5.388 ± 0.036 |
5.572 ± 0.041 | 7.249 ± 0.037 |
1.402 ± 0.02 | 2.37 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
