
Natronincola ferrireducens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Natronincola
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
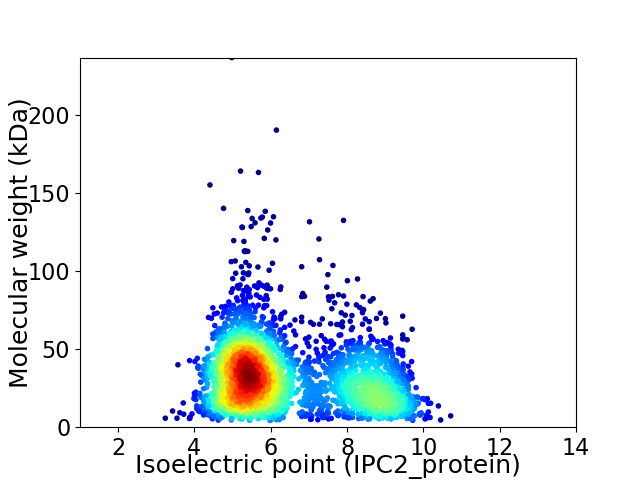
Virtual 2D-PAGE plot for 2920 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8YMF9|A0A1G8YMF9_9CLOT Putative competence-damage inducible protein OS=Natronincola ferrireducens OX=393762 GN=cinA PE=3 SV=1
MM1 pKa = 6.98NHH3 pKa = 6.44WKK5 pKa = 9.83PGQFAKK11 pKa = 10.55ILLALMLIVALLVLTGCGGGTGDD34 pKa = 3.34TTAPSEE40 pKa = 4.13EE41 pKa = 4.33SQEE44 pKa = 4.17EE45 pKa = 4.48VVDD48 pKa = 4.23RR49 pKa = 11.84DD50 pKa = 3.56DD51 pKa = 6.48DD52 pKa = 4.37EE53 pKa = 6.68DD54 pKa = 3.96VDD56 pKa = 4.09RR57 pKa = 11.84KK58 pKa = 10.8KK59 pKa = 11.24GFAHH63 pKa = 7.11FADD66 pKa = 3.99EE67 pKa = 4.47WVVPTIQPPLEE78 pKa = 4.02FVGNDD83 pKa = 3.37GFDD86 pKa = 3.61EE87 pKa = 4.84QILCSTGPTIVTISPEE103 pKa = 3.92GDD105 pKa = 3.09FLRR108 pKa = 11.84RR109 pKa = 11.84IVSSGKK115 pKa = 10.22GYY117 pKa = 10.66YY118 pKa = 9.78DD119 pKa = 3.91YY120 pKa = 11.14PVLSPDD126 pKa = 2.98GTRR129 pKa = 11.84FVVNADD135 pKa = 3.06YY136 pKa = 11.38HH137 pKa = 6.17EE138 pKa = 5.54FIEE141 pKa = 4.56PWEE144 pKa = 3.99MMLLGDD150 pKa = 4.43MDD152 pKa = 3.94TGEE155 pKa = 4.89LIEE158 pKa = 5.36LEE160 pKa = 4.22AAGDD164 pKa = 3.9YY165 pKa = 10.79YY166 pKa = 11.26YY167 pKa = 11.52YY168 pKa = 9.79PASWNHH174 pKa = 6.85DD175 pKa = 3.66GTKK178 pKa = 10.32VIYY181 pKa = 8.81TAYY184 pKa = 10.74DD185 pKa = 3.75EE186 pKa = 5.66DD187 pKa = 3.9WLANIYY193 pKa = 10.29YY194 pKa = 10.22VDD196 pKa = 3.87VDD198 pKa = 3.83TMEE201 pKa = 4.49SGIFLDD207 pKa = 5.0LAEE210 pKa = 4.8LPIDD214 pKa = 3.61RR215 pKa = 11.84EE216 pKa = 4.32VVEE219 pKa = 4.44TAKK222 pKa = 10.75LHH224 pKa = 5.46PNKK227 pKa = 10.5NILYY231 pKa = 9.39FIALPEE237 pKa = 4.29GEE239 pKa = 4.21EE240 pKa = 4.28DD241 pKa = 3.81YY242 pKa = 11.67GVFKK246 pKa = 11.19YY247 pKa = 10.91DD248 pKa = 3.29LDD250 pKa = 4.15EE251 pKa = 4.87GVLEE255 pKa = 4.32QVWTTEE261 pKa = 3.53EE262 pKa = 4.47EE263 pKa = 4.44YY264 pKa = 11.14VYY266 pKa = 10.48DD267 pKa = 3.56IAISPRR273 pKa = 11.84DD274 pKa = 3.72DD275 pKa = 4.07LLAISAGMGITEE287 pKa = 4.58EE288 pKa = 4.62DD289 pKa = 3.86MSTMGLYY296 pKa = 10.32LVYY299 pKa = 10.49PEE301 pKa = 4.4EE302 pKa = 4.59NKK304 pKa = 10.93DD305 pKa = 3.83EE306 pKa = 4.27LLFWEE311 pKa = 5.18EE312 pKa = 4.06DD313 pKa = 3.56VQPIGVSFSPCGKK326 pKa = 10.07YY327 pKa = 10.19LAFDD331 pKa = 4.14ADD333 pKa = 3.61TRR335 pKa = 11.84VIEE338 pKa = 4.41NTWSIYY344 pKa = 10.3IYY346 pKa = 10.82DD347 pKa = 3.43VDD349 pKa = 3.97GRR351 pKa = 11.84DD352 pKa = 3.2GKK354 pKa = 11.25LITRR358 pKa = 11.84YY359 pKa = 9.78IYY361 pKa = 10.08DD362 pKa = 3.3SHH364 pKa = 8.39IEE366 pKa = 4.02PFSPSWGIIPP376 pKa = 4.78
MM1 pKa = 6.98NHH3 pKa = 6.44WKK5 pKa = 9.83PGQFAKK11 pKa = 10.55ILLALMLIVALLVLTGCGGGTGDD34 pKa = 3.34TTAPSEE40 pKa = 4.13EE41 pKa = 4.33SQEE44 pKa = 4.17EE45 pKa = 4.48VVDD48 pKa = 4.23RR49 pKa = 11.84DD50 pKa = 3.56DD51 pKa = 6.48DD52 pKa = 4.37EE53 pKa = 6.68DD54 pKa = 3.96VDD56 pKa = 4.09RR57 pKa = 11.84KK58 pKa = 10.8KK59 pKa = 11.24GFAHH63 pKa = 7.11FADD66 pKa = 3.99EE67 pKa = 4.47WVVPTIQPPLEE78 pKa = 4.02FVGNDD83 pKa = 3.37GFDD86 pKa = 3.61EE87 pKa = 4.84QILCSTGPTIVTISPEE103 pKa = 3.92GDD105 pKa = 3.09FLRR108 pKa = 11.84RR109 pKa = 11.84IVSSGKK115 pKa = 10.22GYY117 pKa = 10.66YY118 pKa = 9.78DD119 pKa = 3.91YY120 pKa = 11.14PVLSPDD126 pKa = 2.98GTRR129 pKa = 11.84FVVNADD135 pKa = 3.06YY136 pKa = 11.38HH137 pKa = 6.17EE138 pKa = 5.54FIEE141 pKa = 4.56PWEE144 pKa = 3.99MMLLGDD150 pKa = 4.43MDD152 pKa = 3.94TGEE155 pKa = 4.89LIEE158 pKa = 5.36LEE160 pKa = 4.22AAGDD164 pKa = 3.9YY165 pKa = 10.79YY166 pKa = 11.26YY167 pKa = 11.52YY168 pKa = 9.79PASWNHH174 pKa = 6.85DD175 pKa = 3.66GTKK178 pKa = 10.32VIYY181 pKa = 8.81TAYY184 pKa = 10.74DD185 pKa = 3.75EE186 pKa = 5.66DD187 pKa = 3.9WLANIYY193 pKa = 10.29YY194 pKa = 10.22VDD196 pKa = 3.87VDD198 pKa = 3.83TMEE201 pKa = 4.49SGIFLDD207 pKa = 5.0LAEE210 pKa = 4.8LPIDD214 pKa = 3.61RR215 pKa = 11.84EE216 pKa = 4.32VVEE219 pKa = 4.44TAKK222 pKa = 10.75LHH224 pKa = 5.46PNKK227 pKa = 10.5NILYY231 pKa = 9.39FIALPEE237 pKa = 4.29GEE239 pKa = 4.21EE240 pKa = 4.28DD241 pKa = 3.81YY242 pKa = 11.67GVFKK246 pKa = 11.19YY247 pKa = 10.91DD248 pKa = 3.29LDD250 pKa = 4.15EE251 pKa = 4.87GVLEE255 pKa = 4.32QVWTTEE261 pKa = 3.53EE262 pKa = 4.47EE263 pKa = 4.44YY264 pKa = 11.14VYY266 pKa = 10.48DD267 pKa = 3.56IAISPRR273 pKa = 11.84DD274 pKa = 3.72DD275 pKa = 4.07LLAISAGMGITEE287 pKa = 4.58EE288 pKa = 4.62DD289 pKa = 3.86MSTMGLYY296 pKa = 10.32LVYY299 pKa = 10.49PEE301 pKa = 4.4EE302 pKa = 4.59NKK304 pKa = 10.93DD305 pKa = 3.83EE306 pKa = 4.27LLFWEE311 pKa = 5.18EE312 pKa = 4.06DD313 pKa = 3.56VQPIGVSFSPCGKK326 pKa = 10.07YY327 pKa = 10.19LAFDD331 pKa = 4.14ADD333 pKa = 3.61TRR335 pKa = 11.84VIEE338 pKa = 4.41NTWSIYY344 pKa = 10.3IYY346 pKa = 10.82DD347 pKa = 3.43VDD349 pKa = 3.97GRR351 pKa = 11.84DD352 pKa = 3.2GKK354 pKa = 11.25LITRR358 pKa = 11.84YY359 pKa = 9.78IYY361 pKa = 10.08DD362 pKa = 3.3SHH364 pKa = 8.39IEE366 pKa = 4.02PFSPSWGIIPP376 pKa = 4.78
Molecular weight: 42.62 kDa
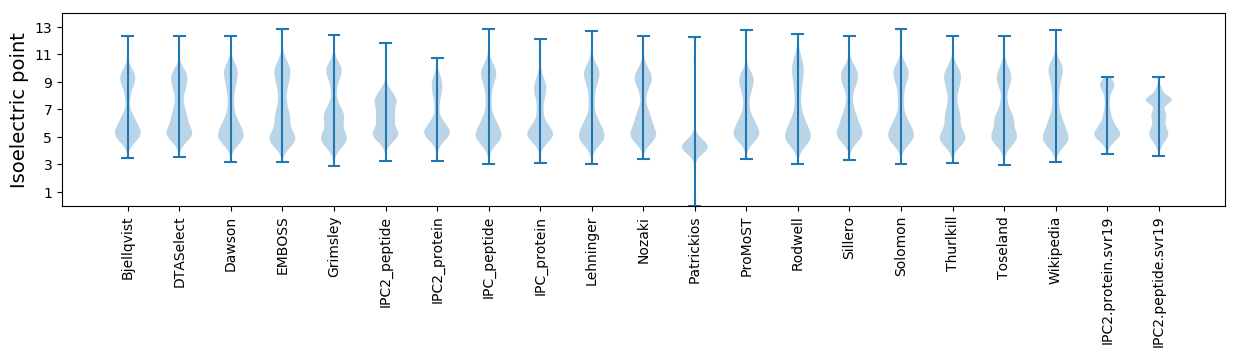
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9C1A1|A0A1G9C1A1_9CLOT Flagellar operon protein OS=Natronincola ferrireducens OX=393762 GN=SAMN05660472_01330 PE=4 SV=1
MM1 pKa = 7.61ARR3 pKa = 11.84VCDD6 pKa = 3.66VCEE9 pKa = 3.97KK10 pKa = 10.85GKK12 pKa = 11.13VYY14 pKa = 10.67GNQVSHH20 pKa = 6.63SNRR23 pKa = 11.84HH24 pKa = 5.35SKK26 pKa = 8.87RR27 pKa = 11.84TWLPNLRR34 pKa = 11.84RR35 pKa = 11.84VKK37 pKa = 10.58AIIDD41 pKa = 3.74GTPKK45 pKa = 10.52RR46 pKa = 11.84INICTRR52 pKa = 11.84CLRR55 pKa = 11.84SGKK58 pKa = 7.94VEE60 pKa = 3.89RR61 pKa = 11.84AII63 pKa = 4.51
MM1 pKa = 7.61ARR3 pKa = 11.84VCDD6 pKa = 3.66VCEE9 pKa = 3.97KK10 pKa = 10.85GKK12 pKa = 11.13VYY14 pKa = 10.67GNQVSHH20 pKa = 6.63SNRR23 pKa = 11.84HH24 pKa = 5.35SKK26 pKa = 8.87RR27 pKa = 11.84TWLPNLRR34 pKa = 11.84RR35 pKa = 11.84VKK37 pKa = 10.58AIIDD41 pKa = 3.74GTPKK45 pKa = 10.52RR46 pKa = 11.84INICTRR52 pKa = 11.84CLRR55 pKa = 11.84SGKK58 pKa = 7.94VEE60 pKa = 3.89RR61 pKa = 11.84AII63 pKa = 4.51
Molecular weight: 7.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
887233 |
40 |
2126 |
303.8 |
34.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.386 ± 0.048 | 0.942 ± 0.016 |
5.104 ± 0.035 | 7.764 ± 0.051 |
4.086 ± 0.037 | 7.115 ± 0.05 |
1.799 ± 0.019 | 9.692 ± 0.043 |
7.842 ± 0.044 | 9.477 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.825 ± 0.021 | 4.958 ± 0.037 |
3.256 ± 0.02 | 3.099 ± 0.028 |
3.837 ± 0.032 | 5.263 ± 0.032 |
5.267 ± 0.027 | 6.789 ± 0.038 |
0.71 ± 0.014 | 3.789 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
