
Rice gall dwarf virus (RGDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Phytoreovirus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
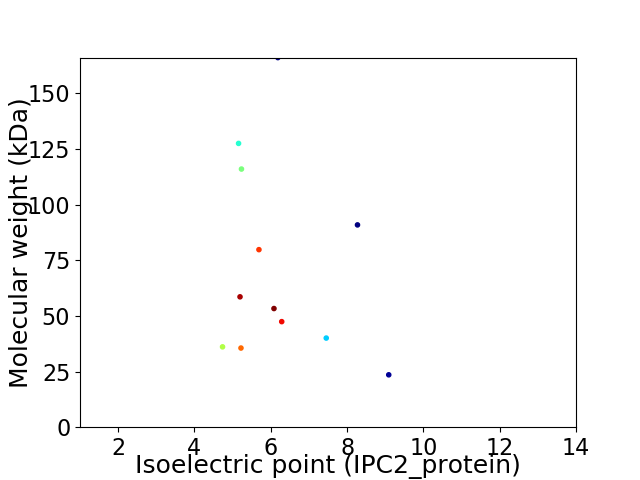
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
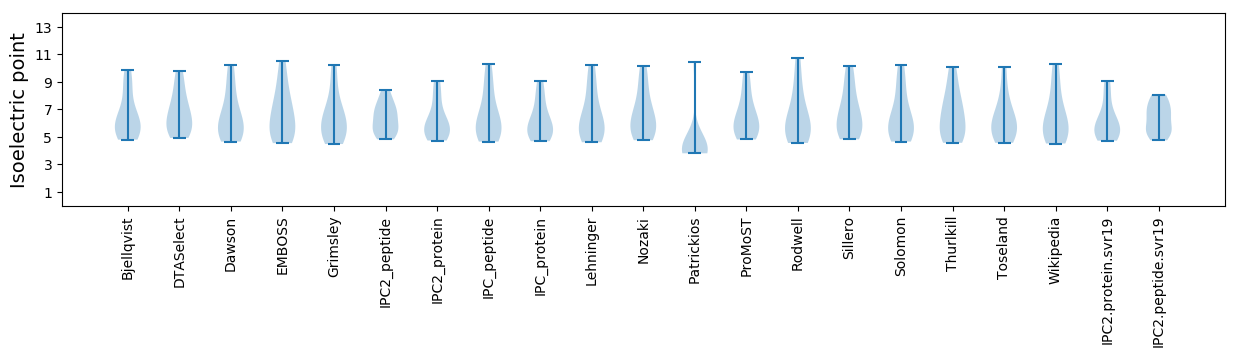
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|Q1WNZ7|MCE_RGDV Putative mRNA-capping enzyme P5 OS=Rice gall dwarf virus OX=10986 PE=3 SV=1
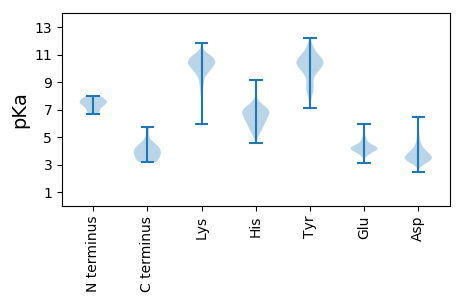
MM1 pKa = 7.68AGKK4 pKa = 9.4LQDD7 pKa = 3.37GVAIAKK13 pKa = 9.87IKK15 pKa = 9.0EE16 pKa = 4.39TINLFCEE23 pKa = 4.4YY24 pKa = 11.03SFGDD28 pKa = 3.47LVNNRR33 pKa = 11.84RR34 pKa = 11.84EE35 pKa = 4.06IVGRR39 pKa = 11.84VHH41 pKa = 7.93DD42 pKa = 4.23ARR44 pKa = 11.84KK45 pKa = 9.27NAALAWPDD53 pKa = 4.76LIMNCFLHH61 pKa = 6.5SASHH65 pKa = 5.48YY66 pKa = 10.24GVVKK70 pKa = 10.28FLLDD74 pKa = 3.13IALSTRR80 pKa = 11.84FGDD83 pKa = 3.85FTLLGVSSQNYY94 pKa = 8.26PFYY97 pKa = 10.58DD98 pKa = 3.34LHH100 pKa = 8.66VVMTKK105 pKa = 10.51AFCNLDD111 pKa = 3.63FAKK114 pKa = 10.71DD115 pKa = 3.79EE116 pKa = 4.09YY117 pKa = 12.0LMINDD122 pKa = 4.62SFSSMMSAFLDD133 pKa = 3.75EE134 pKa = 5.04EE135 pKa = 5.08GVHH138 pKa = 6.0SAMSMEE144 pKa = 4.67LGIHH148 pKa = 6.75DD149 pKa = 3.66IEE151 pKa = 4.71DD152 pKa = 3.51RR153 pKa = 11.84FVLRR157 pKa = 11.84TKK159 pKa = 10.08RR160 pKa = 11.84LFYY163 pKa = 10.37IIHH166 pKa = 7.02EE167 pKa = 4.29YY168 pKa = 10.78HH169 pKa = 6.16MSLDD173 pKa = 4.21EE174 pKa = 4.24IEE176 pKa = 4.48PWLEE180 pKa = 3.88KK181 pKa = 11.03LPDD184 pKa = 3.74ASGGTLLNQKK194 pKa = 9.78SKK196 pKa = 10.54EE197 pKa = 3.83QMRR200 pKa = 11.84VIFSNAKK207 pKa = 9.12VRR209 pKa = 11.84IANSINLYY217 pKa = 8.55VTTHH221 pKa = 5.37TNSYY225 pKa = 9.2NEE227 pKa = 4.13YY228 pKa = 8.09VRR230 pKa = 11.84EE231 pKa = 4.0VAEE234 pKa = 4.18YY235 pKa = 10.86VADD238 pKa = 3.96LWNIQTTTNTQGHH251 pKa = 5.69EE252 pKa = 4.12NEE254 pKa = 4.59LAAEE258 pKa = 4.26DD259 pKa = 4.42FGVLASSSQMNGTKK273 pKa = 9.92SEE275 pKa = 4.59LGDD278 pKa = 3.92SVIKK282 pKa = 10.82SDD284 pKa = 3.57GNEE287 pKa = 4.02VKK289 pKa = 10.75LEE291 pKa = 3.8PAMFTRR297 pKa = 11.84NDD299 pKa = 3.85DD300 pKa = 3.9EE301 pKa = 4.87EE302 pKa = 4.27EE303 pKa = 4.07LAGSEE308 pKa = 4.33FTSLLSDD315 pKa = 4.2DD316 pKa = 4.24GRR318 pKa = 11.84MGG320 pKa = 3.35
MM1 pKa = 7.68AGKK4 pKa = 9.4LQDD7 pKa = 3.37GVAIAKK13 pKa = 9.87IKK15 pKa = 9.0EE16 pKa = 4.39TINLFCEE23 pKa = 4.4YY24 pKa = 11.03SFGDD28 pKa = 3.47LVNNRR33 pKa = 11.84RR34 pKa = 11.84EE35 pKa = 4.06IVGRR39 pKa = 11.84VHH41 pKa = 7.93DD42 pKa = 4.23ARR44 pKa = 11.84KK45 pKa = 9.27NAALAWPDD53 pKa = 4.76LIMNCFLHH61 pKa = 6.5SASHH65 pKa = 5.48YY66 pKa = 10.24GVVKK70 pKa = 10.28FLLDD74 pKa = 3.13IALSTRR80 pKa = 11.84FGDD83 pKa = 3.85FTLLGVSSQNYY94 pKa = 8.26PFYY97 pKa = 10.58DD98 pKa = 3.34LHH100 pKa = 8.66VVMTKK105 pKa = 10.51AFCNLDD111 pKa = 3.63FAKK114 pKa = 10.71DD115 pKa = 3.79EE116 pKa = 4.09YY117 pKa = 12.0LMINDD122 pKa = 4.62SFSSMMSAFLDD133 pKa = 3.75EE134 pKa = 5.04EE135 pKa = 5.08GVHH138 pKa = 6.0SAMSMEE144 pKa = 4.67LGIHH148 pKa = 6.75DD149 pKa = 3.66IEE151 pKa = 4.71DD152 pKa = 3.51RR153 pKa = 11.84FVLRR157 pKa = 11.84TKK159 pKa = 10.08RR160 pKa = 11.84LFYY163 pKa = 10.37IIHH166 pKa = 7.02EE167 pKa = 4.29YY168 pKa = 10.78HH169 pKa = 6.16MSLDD173 pKa = 4.21EE174 pKa = 4.24IEE176 pKa = 4.48PWLEE180 pKa = 3.88KK181 pKa = 11.03LPDD184 pKa = 3.74ASGGTLLNQKK194 pKa = 9.78SKK196 pKa = 10.54EE197 pKa = 3.83QMRR200 pKa = 11.84VIFSNAKK207 pKa = 9.12VRR209 pKa = 11.84IANSINLYY217 pKa = 8.55VTTHH221 pKa = 5.37TNSYY225 pKa = 9.2NEE227 pKa = 4.13YY228 pKa = 8.09VRR230 pKa = 11.84EE231 pKa = 4.0VAEE234 pKa = 4.18YY235 pKa = 10.86VADD238 pKa = 3.96LWNIQTTTNTQGHH251 pKa = 5.69EE252 pKa = 4.12NEE254 pKa = 4.59LAAEE258 pKa = 4.26DD259 pKa = 4.42FGVLASSSQMNGTKK273 pKa = 9.92SEE275 pKa = 4.59LGDD278 pKa = 3.92SVIKK282 pKa = 10.82SDD284 pKa = 3.57GNEE287 pKa = 4.02VKK289 pKa = 10.75LEE291 pKa = 3.8PAMFTRR297 pKa = 11.84NDD299 pKa = 3.85DD300 pKa = 3.9EE301 pKa = 4.87EE302 pKa = 4.27EE303 pKa = 4.07LAGSEE308 pKa = 4.33FTSLLSDD315 pKa = 4.2DD316 pKa = 4.24GRR318 pKa = 11.84MGG320 pKa = 3.35
Molecular weight: 36.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O56834|P2_RGDV Minor outer capsid protein P2 OS=Rice gall dwarf virus OX=10986 PE=1 SV=1
MM1 pKa = 7.42TSNEE5 pKa = 4.15EE6 pKa = 4.11NPVGVQTLKK15 pKa = 10.78PNNDD19 pKa = 3.25AKK21 pKa = 10.72RR22 pKa = 11.84VFEE25 pKa = 3.9MRR27 pKa = 11.84KK28 pKa = 8.77IPSSNVMRR36 pKa = 11.84VEE38 pKa = 3.78YY39 pKa = 9.74TKK41 pKa = 9.85QSKK44 pKa = 10.06YY45 pKa = 8.15RR46 pKa = 11.84TCVCPSALHH55 pKa = 5.94FAEE58 pKa = 4.9EE59 pKa = 4.62CPSNDD64 pKa = 2.93VLFSVGAHH72 pKa = 5.84RR73 pKa = 11.84NMLCVAKK80 pKa = 10.67AFDD83 pKa = 3.57QTKK86 pKa = 9.21PRR88 pKa = 11.84GMQRR92 pKa = 11.84GNRR95 pKa = 11.84RR96 pKa = 11.84NNVAKK101 pKa = 10.84VMTLNEE107 pKa = 4.95LMPKK111 pKa = 10.21DD112 pKa = 4.4DD113 pKa = 4.23SNDD116 pKa = 3.2RR117 pKa = 11.84KK118 pKa = 10.23KK119 pKa = 11.1AKK121 pKa = 9.03TSKK124 pKa = 10.11DD125 pKa = 3.12RR126 pKa = 11.84KK127 pKa = 9.03VEE129 pKa = 3.74KK130 pKa = 10.12SSRR133 pKa = 11.84GRR135 pKa = 11.84ASSSRR140 pKa = 11.84SRR142 pKa = 11.84GRR144 pKa = 11.84GGFSRR149 pKa = 11.84NGRR152 pKa = 11.84DD153 pKa = 3.23RR154 pKa = 11.84RR155 pKa = 11.84EE156 pKa = 3.69TDD158 pKa = 3.16SQDD161 pKa = 3.41EE162 pKa = 4.23YY163 pKa = 11.87SEE165 pKa = 4.6DD166 pKa = 3.21NSYY169 pKa = 11.2KK170 pKa = 10.29SEE172 pKa = 4.09EE173 pKa = 4.27EE174 pKa = 4.01STEE177 pKa = 4.69DD178 pKa = 2.98EE179 pKa = 4.35TAPLTKK185 pKa = 9.76KK186 pKa = 10.79DD187 pKa = 3.91MLKK190 pKa = 9.75MMSMFQSGAKK200 pKa = 9.26SKK202 pKa = 10.48RR203 pKa = 11.84RR204 pKa = 11.84PRR206 pKa = 3.77
MM1 pKa = 7.42TSNEE5 pKa = 4.15EE6 pKa = 4.11NPVGVQTLKK15 pKa = 10.78PNNDD19 pKa = 3.25AKK21 pKa = 10.72RR22 pKa = 11.84VFEE25 pKa = 3.9MRR27 pKa = 11.84KK28 pKa = 8.77IPSSNVMRR36 pKa = 11.84VEE38 pKa = 3.78YY39 pKa = 9.74TKK41 pKa = 9.85QSKK44 pKa = 10.06YY45 pKa = 8.15RR46 pKa = 11.84TCVCPSALHH55 pKa = 5.94FAEE58 pKa = 4.9EE59 pKa = 4.62CPSNDD64 pKa = 2.93VLFSVGAHH72 pKa = 5.84RR73 pKa = 11.84NMLCVAKK80 pKa = 10.67AFDD83 pKa = 3.57QTKK86 pKa = 9.21PRR88 pKa = 11.84GMQRR92 pKa = 11.84GNRR95 pKa = 11.84RR96 pKa = 11.84NNVAKK101 pKa = 10.84VMTLNEE107 pKa = 4.95LMPKK111 pKa = 10.21DD112 pKa = 4.4DD113 pKa = 4.23SNDD116 pKa = 3.2RR117 pKa = 11.84KK118 pKa = 10.23KK119 pKa = 11.1AKK121 pKa = 9.03TSKK124 pKa = 10.11DD125 pKa = 3.12RR126 pKa = 11.84KK127 pKa = 9.03VEE129 pKa = 3.74KK130 pKa = 10.12SSRR133 pKa = 11.84GRR135 pKa = 11.84ASSSRR140 pKa = 11.84SRR142 pKa = 11.84GRR144 pKa = 11.84GGFSRR149 pKa = 11.84NGRR152 pKa = 11.84DD153 pKa = 3.23RR154 pKa = 11.84RR155 pKa = 11.84EE156 pKa = 3.69TDD158 pKa = 3.16SQDD161 pKa = 3.41EE162 pKa = 4.23YY163 pKa = 11.87SEE165 pKa = 4.6DD166 pKa = 3.21NSYY169 pKa = 11.2KK170 pKa = 10.29SEE172 pKa = 4.09EE173 pKa = 4.27EE174 pKa = 4.01STEE177 pKa = 4.69DD178 pKa = 2.98EE179 pKa = 4.35TAPLTKK185 pKa = 9.76KK186 pKa = 10.79DD187 pKa = 3.91MLKK190 pKa = 9.75MMSMFQSGAKK200 pKa = 9.26SKK202 pKa = 10.48RR203 pKa = 11.84RR204 pKa = 11.84PRR206 pKa = 3.77
Molecular weight: 23.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7782 |
206 |
1458 |
648.5 |
72.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.808 ± 0.306 | 1.079 ± 0.096 |
6.181 ± 0.324 | 5.423 ± 0.392 |
3.765 ± 0.252 | 5.217 ± 0.375 |
1.748 ± 0.163 | 7.466 ± 0.359 |
5.795 ± 0.469 | 8.314 ± 0.404 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.624 ± 0.169 | 6.412 ± 0.252 |
3.958 ± 0.317 | 2.917 ± 0.252 |
4.87 ± 0.369 | 9.098 ± 0.275 |
6.207 ± 0.308 | 7.183 ± 0.398 |
0.784 ± 0.097 | 4.151 ± 0.305 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
