
Frigoribacterium sp. Leaf172
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Frigoribacterium; unclassified Frigoribacterium
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
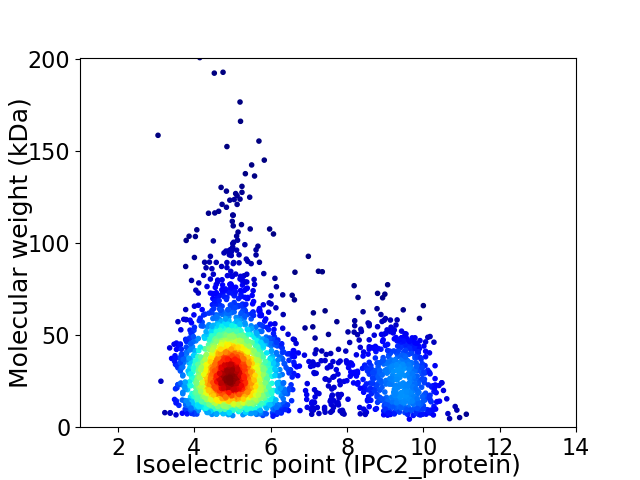
Virtual 2D-PAGE plot for 2858 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5N8R6|A0A0Q5N8R6_9MICO Uncharacterized protein OS=Frigoribacterium sp. Leaf172 OX=1736285 GN=ASF89_01875 PE=4 SV=1
MM1 pKa = 7.15GAAAAASLALVLTGCSAGGSNSSSTPSDD29 pKa = 3.19TLVVYY34 pKa = 8.99TGQAGDD40 pKa = 3.56YY41 pKa = 9.88QINFNPYY48 pKa = 8.18SPSSIGGIGAIYY60 pKa = 10.15EE61 pKa = 4.3SLFFVTNVNTEE72 pKa = 3.67AYY74 pKa = 8.31EE75 pKa = 3.96PLLGTEE81 pKa = 4.36YY82 pKa = 9.84TWNDD86 pKa = 3.26AGTEE90 pKa = 4.01LAVTLRR96 pKa = 11.84DD97 pKa = 3.53DD98 pKa = 4.01ATWSDD103 pKa = 4.29GEE105 pKa = 4.82PFTADD110 pKa = 2.79DD111 pKa = 4.06VVFTFQMLLDD121 pKa = 4.12TPSLNTSGFDD131 pKa = 3.82GAVTAPDD138 pKa = 3.51DD139 pKa = 3.71THH141 pKa = 8.99VVFTWDD147 pKa = 2.94EE148 pKa = 4.01PAFVAGPTLLGRR160 pKa = 11.84TPIVPEE166 pKa = 4.32HH167 pKa = 6.34LWSGIDD173 pKa = 3.34PTTDD177 pKa = 2.75VMAEE181 pKa = 4.01PVGTGAFTLEE191 pKa = 4.09NFKK194 pKa = 11.11AQAFTLASNPEE205 pKa = 3.98YY206 pKa = 10.09WGGEE210 pKa = 3.96PEE212 pKa = 4.42VKK214 pKa = 9.92KK215 pKa = 11.02VRR217 pKa = 11.84YY218 pKa = 9.31LSLSGNTAGADD229 pKa = 3.23ALAAGTIDD237 pKa = 3.65WQTGPVPDD245 pKa = 4.28MEE247 pKa = 4.51NVSEE251 pKa = 4.45NYY253 pKa = 9.45PGYY256 pKa = 10.56DD257 pKa = 3.44AVTIPQNQVALLTCSSVEE275 pKa = 4.16LGCEE279 pKa = 4.6GPQTDD284 pKa = 3.7PAVRR288 pKa = 11.84QAIYY292 pKa = 10.01HH293 pKa = 6.15ALNRR297 pKa = 11.84DD298 pKa = 3.56QVNSLAFQNTASEE311 pKa = 4.12ISPTFALTSTQEE323 pKa = 4.1DD324 pKa = 4.85LISADD329 pKa = 3.16VAEE332 pKa = 4.98PVAPSAPDD340 pKa = 3.46LDD342 pKa = 4.16KK343 pKa = 11.36SAEE346 pKa = 4.04LLEE349 pKa = 4.53GAGYY353 pKa = 8.36TKK355 pKa = 10.9GGDD358 pKa = 3.16GVYY361 pKa = 10.69AKK363 pKa = 10.68DD364 pKa = 3.91GEE366 pKa = 4.33PLKK369 pKa = 10.59LTVEE373 pKa = 4.65VVTGWTDD380 pKa = 3.58YY381 pKa = 9.72ITAIDD386 pKa = 3.96TMGQQLKK393 pKa = 10.41AAGIEE398 pKa = 4.15LTASQSSWNEE408 pKa = 3.51WTDD411 pKa = 3.27KK412 pKa = 10.66KK413 pKa = 11.42SKK415 pKa = 10.77GNYY418 pKa = 8.5EE419 pKa = 3.96LAIDD423 pKa = 4.46SLGQGPASDD432 pKa = 4.07PFYY435 pKa = 11.08LYY437 pKa = 10.62DD438 pKa = 3.35QYY440 pKa = 11.93FNTAYY445 pKa = 8.97TAEE448 pKa = 4.27VGEE451 pKa = 4.22AAPTNMARR459 pKa = 11.84FSDD462 pKa = 4.21PAVDD466 pKa = 3.65AALAEE471 pKa = 4.4LKK473 pKa = 10.78AVDD476 pKa = 4.32PADD479 pKa = 3.47TAARR483 pKa = 11.84QAQFDD488 pKa = 4.37VIQAAIVEE496 pKa = 4.47DD497 pKa = 3.91MPYY500 pKa = 10.07IPVMTGGTTSEE511 pKa = 4.01FHH513 pKa = 6.61SSKK516 pKa = 9.43FTGWPTMDD524 pKa = 4.96DD525 pKa = 4.17LYY527 pKa = 11.3AFPAIWASPDD537 pKa = 3.04NAQVFKK543 pKa = 10.84NLKK546 pKa = 7.53PTGEE550 pKa = 3.98
MM1 pKa = 7.15GAAAAASLALVLTGCSAGGSNSSSTPSDD29 pKa = 3.19TLVVYY34 pKa = 8.99TGQAGDD40 pKa = 3.56YY41 pKa = 9.88QINFNPYY48 pKa = 8.18SPSSIGGIGAIYY60 pKa = 10.15EE61 pKa = 4.3SLFFVTNVNTEE72 pKa = 3.67AYY74 pKa = 8.31EE75 pKa = 3.96PLLGTEE81 pKa = 4.36YY82 pKa = 9.84TWNDD86 pKa = 3.26AGTEE90 pKa = 4.01LAVTLRR96 pKa = 11.84DD97 pKa = 3.53DD98 pKa = 4.01ATWSDD103 pKa = 4.29GEE105 pKa = 4.82PFTADD110 pKa = 2.79DD111 pKa = 4.06VVFTFQMLLDD121 pKa = 4.12TPSLNTSGFDD131 pKa = 3.82GAVTAPDD138 pKa = 3.51DD139 pKa = 3.71THH141 pKa = 8.99VVFTWDD147 pKa = 2.94EE148 pKa = 4.01PAFVAGPTLLGRR160 pKa = 11.84TPIVPEE166 pKa = 4.32HH167 pKa = 6.34LWSGIDD173 pKa = 3.34PTTDD177 pKa = 2.75VMAEE181 pKa = 4.01PVGTGAFTLEE191 pKa = 4.09NFKK194 pKa = 11.11AQAFTLASNPEE205 pKa = 3.98YY206 pKa = 10.09WGGEE210 pKa = 3.96PEE212 pKa = 4.42VKK214 pKa = 9.92KK215 pKa = 11.02VRR217 pKa = 11.84YY218 pKa = 9.31LSLSGNTAGADD229 pKa = 3.23ALAAGTIDD237 pKa = 3.65WQTGPVPDD245 pKa = 4.28MEE247 pKa = 4.51NVSEE251 pKa = 4.45NYY253 pKa = 9.45PGYY256 pKa = 10.56DD257 pKa = 3.44AVTIPQNQVALLTCSSVEE275 pKa = 4.16LGCEE279 pKa = 4.6GPQTDD284 pKa = 3.7PAVRR288 pKa = 11.84QAIYY292 pKa = 10.01HH293 pKa = 6.15ALNRR297 pKa = 11.84DD298 pKa = 3.56QVNSLAFQNTASEE311 pKa = 4.12ISPTFALTSTQEE323 pKa = 4.1DD324 pKa = 4.85LISADD329 pKa = 3.16VAEE332 pKa = 4.98PVAPSAPDD340 pKa = 3.46LDD342 pKa = 4.16KK343 pKa = 11.36SAEE346 pKa = 4.04LLEE349 pKa = 4.53GAGYY353 pKa = 8.36TKK355 pKa = 10.9GGDD358 pKa = 3.16GVYY361 pKa = 10.69AKK363 pKa = 10.68DD364 pKa = 3.91GEE366 pKa = 4.33PLKK369 pKa = 10.59LTVEE373 pKa = 4.65VVTGWTDD380 pKa = 3.58YY381 pKa = 9.72ITAIDD386 pKa = 3.96TMGQQLKK393 pKa = 10.41AAGIEE398 pKa = 4.15LTASQSSWNEE408 pKa = 3.51WTDD411 pKa = 3.27KK412 pKa = 10.66KK413 pKa = 11.42SKK415 pKa = 10.77GNYY418 pKa = 8.5EE419 pKa = 3.96LAIDD423 pKa = 4.46SLGQGPASDD432 pKa = 4.07PFYY435 pKa = 11.08LYY437 pKa = 10.62DD438 pKa = 3.35QYY440 pKa = 11.93FNTAYY445 pKa = 8.97TAEE448 pKa = 4.27VGEE451 pKa = 4.22AAPTNMARR459 pKa = 11.84FSDD462 pKa = 4.21PAVDD466 pKa = 3.65AALAEE471 pKa = 4.4LKK473 pKa = 10.78AVDD476 pKa = 4.32PADD479 pKa = 3.47TAARR483 pKa = 11.84QAQFDD488 pKa = 4.37VIQAAIVEE496 pKa = 4.47DD497 pKa = 3.91MPYY500 pKa = 10.07IPVMTGGTTSEE511 pKa = 4.01FHH513 pKa = 6.61SSKK516 pKa = 9.43FTGWPTMDD524 pKa = 4.96DD525 pKa = 4.17LYY527 pKa = 11.3AFPAIWASPDD537 pKa = 3.04NAQVFKK543 pKa = 10.84NLKK546 pKa = 7.53PTGEE550 pKa = 3.98
Molecular weight: 58.36 kDa
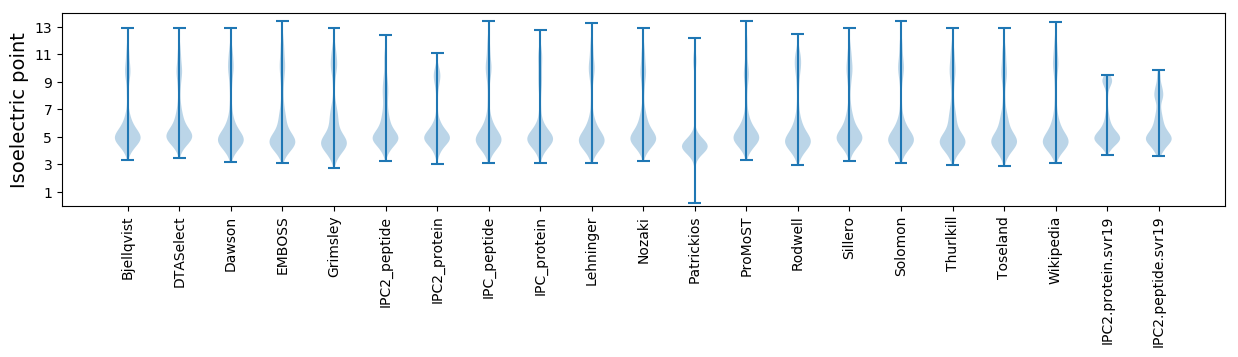
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5N197|A0A0Q5N197_9MICO Epimerase domain-containing protein OS=Frigoribacterium sp. Leaf172 OX=1736285 GN=ASF89_04105 PE=4 SV=1
MM1 pKa = 7.01TRR3 pKa = 11.84RR4 pKa = 11.84GGAALRR10 pKa = 11.84RR11 pKa = 11.84GQLAVSKK18 pKa = 10.65VLAFVSALALLVGLVRR34 pKa = 11.84ILQAHH39 pKa = 5.43VVAGAAFVIIAVALIAVAFALARR62 pKa = 11.84AATRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 3.56
MM1 pKa = 7.01TRR3 pKa = 11.84RR4 pKa = 11.84GGAALRR10 pKa = 11.84RR11 pKa = 11.84GQLAVSKK18 pKa = 10.65VLAFVSALALLVGLVRR34 pKa = 11.84ILQAHH39 pKa = 5.43VVAGAAFVIIAVALIAVAFALARR62 pKa = 11.84AATRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 3.56
Molecular weight: 7.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
913754 |
38 |
1957 |
319.7 |
33.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.302 ± 0.056 | 0.425 ± 0.009 |
6.771 ± 0.051 | 5.39 ± 0.044 |
2.975 ± 0.032 | 9.266 ± 0.045 |
1.885 ± 0.023 | 3.95 ± 0.033 |
1.734 ± 0.034 | 10.048 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.625 ± 0.017 | 1.691 ± 0.021 |
5.398 ± 0.036 | 2.549 ± 0.028 |
7.426 ± 0.055 | 6.123 ± 0.032 |
6.412 ± 0.058 | 9.804 ± 0.05 |
1.374 ± 0.021 | 1.851 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
