
Le Dantec virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Le Dantec ledantevirus
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
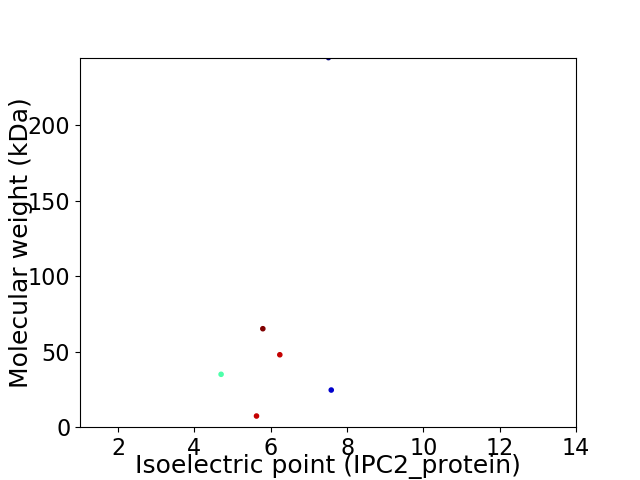
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
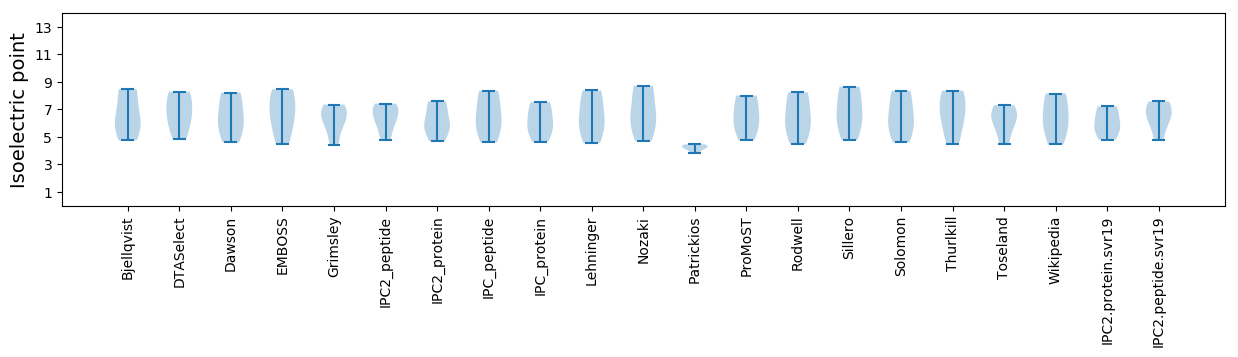
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0D3R1W0|A0A0D3R1W0_9RHAB Phosphoprotein OS=Le Dantec virus OX=318848 PE=4 SV=1
MM1 pKa = 7.48LSSRR5 pKa = 11.84DD6 pKa = 3.4RR7 pKa = 11.84VKK9 pKa = 11.03AIASKK14 pKa = 11.0EE15 pKa = 3.92FFNDD19 pKa = 2.63IDD21 pKa = 3.78GKK23 pKa = 9.94QFEE26 pKa = 4.33EE27 pKa = 4.23DD28 pKa = 3.16QGEE31 pKa = 4.3EE32 pKa = 4.12EE33 pKa = 4.69HH34 pKa = 6.77VSPVPEE40 pKa = 4.48IDD42 pKa = 4.18SDD44 pKa = 4.05FKK46 pKa = 11.21KK47 pKa = 10.96LKK49 pKa = 10.62GSLDD53 pKa = 3.63DD54 pKa = 4.29KK55 pKa = 11.32EE56 pKa = 4.4EE57 pKa = 4.15EE58 pKa = 4.2EE59 pKa = 4.94GFDD62 pKa = 4.14DD63 pKa = 3.47VTEE66 pKa = 4.27IVDD69 pKa = 3.91EE70 pKa = 4.58NGSDD74 pKa = 3.65SDD76 pKa = 4.01SAGDD80 pKa = 3.57INLMISNSEE89 pKa = 3.95NSGDD93 pKa = 4.16DD94 pKa = 3.94DD95 pKa = 4.36NPAALVGRR103 pKa = 11.84HH104 pKa = 5.56EE105 pKa = 4.74YY106 pKa = 10.44IEE108 pKa = 4.3TDD110 pKa = 3.36LKK112 pKa = 11.2HH113 pKa = 6.25FALSDD118 pKa = 3.55EE119 pKa = 4.93LEE121 pKa = 4.29SHH123 pKa = 7.07LSSQEE128 pKa = 3.43EE129 pKa = 4.14IAAHH133 pKa = 5.93EE134 pKa = 4.54SGHH137 pKa = 6.26RR138 pKa = 11.84NSSLNISLPKK148 pKa = 10.37LVLDD152 pKa = 4.9EE153 pKa = 5.38IKK155 pKa = 10.52DD156 pKa = 3.7QKK158 pKa = 11.17EE159 pKa = 3.66LDD161 pKa = 3.97EE162 pKa = 4.49TCMGLLHH169 pKa = 7.1AFAAHH174 pKa = 6.93LGWSVVPMYY183 pKa = 10.84SQVSEE188 pKa = 4.11EE189 pKa = 3.82KK190 pKa = 10.87LIINVKK196 pKa = 6.97QVKK199 pKa = 9.67RR200 pKa = 11.84SKK202 pKa = 9.6QHH204 pKa = 5.28MSISSTDD211 pKa = 2.81MDD213 pKa = 3.68QPVVEE218 pKa = 4.89NLAKK222 pKa = 10.34VNRR225 pKa = 11.84GRR227 pKa = 11.84SSEE230 pKa = 4.03RR231 pKa = 11.84FEE233 pKa = 5.41GKK235 pKa = 9.97DD236 pKa = 3.02KK237 pKa = 11.27EE238 pKa = 4.28RR239 pKa = 11.84FKK241 pKa = 11.48EE242 pKa = 4.51FVEE245 pKa = 4.52KK246 pKa = 10.28MSSGQIRR253 pKa = 11.84FIKK256 pKa = 9.97KK257 pKa = 9.27KK258 pKa = 9.74KK259 pKa = 10.09NGFISLTLATPGVTMEE275 pKa = 4.63LVKK278 pKa = 10.58QALDD282 pKa = 3.32QCVLEE287 pKa = 4.76KK288 pKa = 11.03DD289 pKa = 4.15VIDD292 pKa = 4.19MILRR296 pKa = 11.84EE297 pKa = 4.62SNMLTLVKK305 pKa = 9.13TLCIYY310 pKa = 10.01PP311 pKa = 3.83
MM1 pKa = 7.48LSSRR5 pKa = 11.84DD6 pKa = 3.4RR7 pKa = 11.84VKK9 pKa = 11.03AIASKK14 pKa = 11.0EE15 pKa = 3.92FFNDD19 pKa = 2.63IDD21 pKa = 3.78GKK23 pKa = 9.94QFEE26 pKa = 4.33EE27 pKa = 4.23DD28 pKa = 3.16QGEE31 pKa = 4.3EE32 pKa = 4.12EE33 pKa = 4.69HH34 pKa = 6.77VSPVPEE40 pKa = 4.48IDD42 pKa = 4.18SDD44 pKa = 4.05FKK46 pKa = 11.21KK47 pKa = 10.96LKK49 pKa = 10.62GSLDD53 pKa = 3.63DD54 pKa = 4.29KK55 pKa = 11.32EE56 pKa = 4.4EE57 pKa = 4.15EE58 pKa = 4.2EE59 pKa = 4.94GFDD62 pKa = 4.14DD63 pKa = 3.47VTEE66 pKa = 4.27IVDD69 pKa = 3.91EE70 pKa = 4.58NGSDD74 pKa = 3.65SDD76 pKa = 4.01SAGDD80 pKa = 3.57INLMISNSEE89 pKa = 3.95NSGDD93 pKa = 4.16DD94 pKa = 3.94DD95 pKa = 4.36NPAALVGRR103 pKa = 11.84HH104 pKa = 5.56EE105 pKa = 4.74YY106 pKa = 10.44IEE108 pKa = 4.3TDD110 pKa = 3.36LKK112 pKa = 11.2HH113 pKa = 6.25FALSDD118 pKa = 3.55EE119 pKa = 4.93LEE121 pKa = 4.29SHH123 pKa = 7.07LSSQEE128 pKa = 3.43EE129 pKa = 4.14IAAHH133 pKa = 5.93EE134 pKa = 4.54SGHH137 pKa = 6.26RR138 pKa = 11.84NSSLNISLPKK148 pKa = 10.37LVLDD152 pKa = 4.9EE153 pKa = 5.38IKK155 pKa = 10.52DD156 pKa = 3.7QKK158 pKa = 11.17EE159 pKa = 3.66LDD161 pKa = 3.97EE162 pKa = 4.49TCMGLLHH169 pKa = 7.1AFAAHH174 pKa = 6.93LGWSVVPMYY183 pKa = 10.84SQVSEE188 pKa = 4.11EE189 pKa = 3.82KK190 pKa = 10.87LIINVKK196 pKa = 6.97QVKK199 pKa = 9.67RR200 pKa = 11.84SKK202 pKa = 9.6QHH204 pKa = 5.28MSISSTDD211 pKa = 2.81MDD213 pKa = 3.68QPVVEE218 pKa = 4.89NLAKK222 pKa = 10.34VNRR225 pKa = 11.84GRR227 pKa = 11.84SSEE230 pKa = 4.03RR231 pKa = 11.84FEE233 pKa = 5.41GKK235 pKa = 9.97DD236 pKa = 3.02KK237 pKa = 11.27EE238 pKa = 4.28RR239 pKa = 11.84FKK241 pKa = 11.48EE242 pKa = 4.51FVEE245 pKa = 4.52KK246 pKa = 10.28MSSGQIRR253 pKa = 11.84FIKK256 pKa = 9.97KK257 pKa = 9.27KK258 pKa = 9.74KK259 pKa = 10.09NGFISLTLATPGVTMEE275 pKa = 4.63LVKK278 pKa = 10.58QALDD282 pKa = 3.32QCVLEE287 pKa = 4.76KK288 pKa = 11.03DD289 pKa = 4.15VIDD292 pKa = 4.19MILRR296 pKa = 11.84EE297 pKa = 4.62SNMLTLVKK305 pKa = 9.13TLCIYY310 pKa = 10.01PP311 pKa = 3.83
Molecular weight: 35.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R1R3|A0A0D3R1R3_9RHAB GDP polyribonucleotidyltransferase OS=Le Dantec virus OX=318848 PE=4 SV=1
MM1 pKa = 7.56FSRR4 pKa = 11.84FKK6 pKa = 10.93KK7 pKa = 10.53SLSKK11 pKa = 10.55KK12 pKa = 10.16GKK14 pKa = 9.89NDD16 pKa = 3.9DD17 pKa = 3.65NQLLYY22 pKa = 10.77DD23 pKa = 4.34PPAYY27 pKa = 10.52DD28 pKa = 3.7SDD30 pKa = 5.48DD31 pKa = 3.52FFAPIKK37 pKa = 10.37PSAPDD42 pKa = 3.62LEE44 pKa = 6.12LEE46 pKa = 4.41DD47 pKa = 4.31FRR49 pKa = 11.84KK50 pKa = 8.53EE51 pKa = 4.18TLHH54 pKa = 7.74VEE56 pKa = 3.78AEE58 pKa = 4.56LIIKK62 pKa = 6.98THH64 pKa = 6.22AGIKK68 pKa = 9.23TMEE71 pKa = 4.43EE72 pKa = 3.76LLKK75 pKa = 10.7ILGIWIDD82 pKa = 3.54QCSGPIRR89 pKa = 11.84QRR91 pKa = 11.84HH92 pKa = 5.43LDD94 pKa = 3.33LWVYY98 pKa = 10.84LCLGLHH104 pKa = 5.5VRR106 pKa = 11.84RR107 pKa = 11.84DD108 pKa = 3.82VGIKK112 pKa = 9.91SYY114 pKa = 11.01NVYY117 pKa = 10.16RR118 pKa = 11.84AALDD122 pKa = 3.47SSVVFLHH129 pKa = 7.06APKK132 pKa = 9.16TQGKK136 pKa = 8.16FQFVPCHH143 pKa = 5.4QFFTQKK149 pKa = 10.91YY150 pKa = 8.04KK151 pKa = 11.27GKK153 pKa = 10.22DD154 pKa = 3.11CDD156 pKa = 3.69VDD158 pKa = 4.32FTCKK162 pKa = 10.19FKK164 pKa = 9.64EE165 pKa = 4.51TKK167 pKa = 9.15RR168 pKa = 11.84TGIPAHH174 pKa = 6.28TIYY177 pKa = 11.1NQVLKK182 pKa = 10.96SGFPPPKK189 pKa = 9.17PIEE192 pKa = 4.05VFAGYY197 pKa = 10.61DD198 pKa = 3.26VTVSVNKK205 pKa = 10.49NGEE208 pKa = 3.98HH209 pKa = 6.86EE210 pKa = 4.33IAASATT216 pKa = 3.55
MM1 pKa = 7.56FSRR4 pKa = 11.84FKK6 pKa = 10.93KK7 pKa = 10.53SLSKK11 pKa = 10.55KK12 pKa = 10.16GKK14 pKa = 9.89NDD16 pKa = 3.9DD17 pKa = 3.65NQLLYY22 pKa = 10.77DD23 pKa = 4.34PPAYY27 pKa = 10.52DD28 pKa = 3.7SDD30 pKa = 5.48DD31 pKa = 3.52FFAPIKK37 pKa = 10.37PSAPDD42 pKa = 3.62LEE44 pKa = 6.12LEE46 pKa = 4.41DD47 pKa = 4.31FRR49 pKa = 11.84KK50 pKa = 8.53EE51 pKa = 4.18TLHH54 pKa = 7.74VEE56 pKa = 3.78AEE58 pKa = 4.56LIIKK62 pKa = 6.98THH64 pKa = 6.22AGIKK68 pKa = 9.23TMEE71 pKa = 4.43EE72 pKa = 3.76LLKK75 pKa = 10.7ILGIWIDD82 pKa = 3.54QCSGPIRR89 pKa = 11.84QRR91 pKa = 11.84HH92 pKa = 5.43LDD94 pKa = 3.33LWVYY98 pKa = 10.84LCLGLHH104 pKa = 5.5VRR106 pKa = 11.84RR107 pKa = 11.84DD108 pKa = 3.82VGIKK112 pKa = 9.91SYY114 pKa = 11.01NVYY117 pKa = 10.16RR118 pKa = 11.84AALDD122 pKa = 3.47SSVVFLHH129 pKa = 7.06APKK132 pKa = 9.16TQGKK136 pKa = 8.16FQFVPCHH143 pKa = 5.4QFFTQKK149 pKa = 10.91YY150 pKa = 8.04KK151 pKa = 11.27GKK153 pKa = 10.22DD154 pKa = 3.11CDD156 pKa = 3.69VDD158 pKa = 4.32FTCKK162 pKa = 10.19FKK164 pKa = 9.64EE165 pKa = 4.51TKK167 pKa = 9.15RR168 pKa = 11.84TGIPAHH174 pKa = 6.28TIYY177 pKa = 11.1NQVLKK182 pKa = 10.96SGFPPPKK189 pKa = 9.17PIEE192 pKa = 4.05VFAGYY197 pKa = 10.61DD198 pKa = 3.26VTVSVNKK205 pKa = 10.49NGEE208 pKa = 3.98HH209 pKa = 6.86EE210 pKa = 4.33IAASATT216 pKa = 3.55
Molecular weight: 24.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3714 |
64 |
2126 |
619.0 |
70.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.985 ± 0.778 | 1.939 ± 0.301 |
6.327 ± 0.41 | 6.651 ± 0.739 |
4.793 ± 0.358 | 6.193 ± 0.352 |
2.881 ± 0.286 | 6.893 ± 0.825 |
7.27 ± 0.432 | 9.828 ± 0.663 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.196 | 4.604 ± 0.367 |
4.308 ± 0.258 | 2.8 ± 0.196 |
5.197 ± 0.413 | 7.997 ± 0.367 |
5.089 ± 0.413 | 5.52 ± 0.688 |
1.642 ± 0.217 | 3.339 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
