
Penicillium stoloniferum virus F
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Gammapartitivirus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
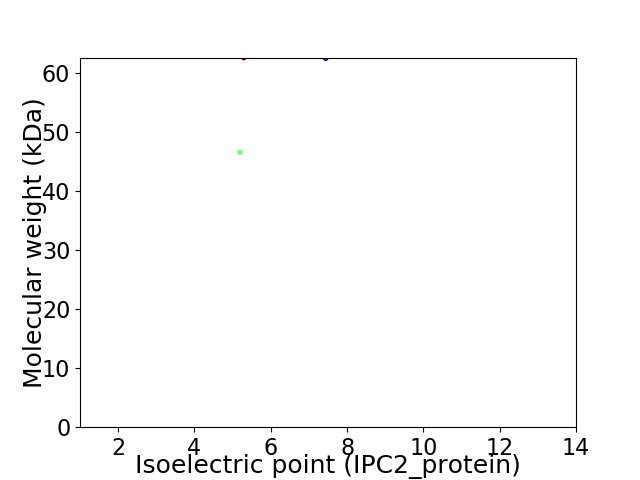
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q4G3H2|Q4G3H2_9VIRU Putative dsRNA-dependent RNA polymerase OS=Penicillium stoloniferum virus F OX=296210 PE=4 SV=1
MM1 pKa = 7.92SFEE4 pKa = 4.24TSEE7 pKa = 4.11GMSRR11 pKa = 11.84PGDD14 pKa = 3.54NPNKK18 pKa = 10.32LNAKK22 pKa = 8.69PRR24 pKa = 11.84QSARR28 pKa = 11.84PKK30 pKa = 9.11TRR32 pKa = 11.84NSTAQSNQTMRR43 pKa = 11.84LGWIDD48 pKa = 4.7PLPQVDD54 pKa = 4.42TIFPLGLEE62 pKa = 4.5PNVEE66 pKa = 4.66SIPAGEE72 pKa = 4.35VEE74 pKa = 5.41LDD76 pKa = 3.53FNLPEE81 pKa = 4.38TIAKK85 pKa = 9.35PFADD89 pKa = 3.58TVTSVGDD96 pKa = 4.28RR97 pKa = 11.84IQLVDD102 pKa = 4.32DD103 pKa = 4.59DD104 pKa = 5.09KK105 pKa = 12.05EE106 pKa = 4.48NIATSIYY113 pKa = 10.37GLSFFKK119 pKa = 10.59AARR122 pKa = 11.84QLYY125 pKa = 8.14STMLDD130 pKa = 3.05HH131 pKa = 7.2EE132 pKa = 4.67KK133 pKa = 10.83AVNQPLKK140 pKa = 10.47AVYY143 pKa = 10.17YY144 pKa = 10.5DD145 pKa = 3.65EE146 pKa = 4.75TPIPAHH152 pKa = 5.97MSGALGIIGHH162 pKa = 6.41MKK164 pKa = 9.66TKK166 pKa = 10.67VGDD169 pKa = 3.93VLVKK173 pKa = 10.72DD174 pKa = 3.67AGVLFKK180 pKa = 10.72RR181 pKa = 11.84GTAAGVTKK189 pKa = 10.62FSEE192 pKa = 3.95IDD194 pKa = 3.64NDD196 pKa = 3.92KK197 pKa = 9.73TWNLDD202 pKa = 3.23CSKK205 pKa = 10.66LVWADD210 pKa = 3.45HH211 pKa = 6.66SSLSMIKK218 pKa = 10.15RR219 pKa = 11.84LASEE223 pKa = 5.13KK224 pKa = 10.18ISQLVKK230 pKa = 9.48QRR232 pKa = 11.84YY233 pKa = 8.49RR234 pKa = 11.84VTDD237 pKa = 3.71AQGHH241 pKa = 5.21VYY243 pKa = 9.78SVSMPQLTDD252 pKa = 3.15QALPDD257 pKa = 4.25YY258 pKa = 9.93YY259 pKa = 11.35DD260 pKa = 4.84SIPDD264 pKa = 3.43VAPNSDD270 pKa = 3.4QLRR273 pKa = 11.84VLTAALQMSLAQFRR287 pKa = 11.84NDD289 pKa = 3.84EE290 pKa = 4.34LPHH293 pKa = 8.28DD294 pKa = 4.55EE295 pKa = 5.32DD296 pKa = 6.08RR297 pKa = 11.84SDD299 pKa = 5.4LLTTLDD305 pKa = 4.31LLYY308 pKa = 10.93ADD310 pKa = 4.42GAYY313 pKa = 9.85EE314 pKa = 4.09ISALRR319 pKa = 11.84DD320 pKa = 3.37QFEE323 pKa = 4.79LLMARR328 pKa = 11.84YY329 pKa = 6.15TTDD332 pKa = 3.63FKK334 pKa = 11.18WRR336 pKa = 11.84VEE338 pKa = 4.52SIFKK342 pKa = 10.43VGPPPAGTTGYY353 pKa = 10.34GAQTVSSTGNTARR366 pKa = 11.84WQFPLSDD373 pKa = 3.42ADD375 pKa = 3.68INIGYY380 pKa = 9.29LFSPSKK386 pKa = 10.74SFSLFPKK393 pKa = 9.24MVGYY397 pKa = 9.88SKK399 pKa = 10.51RR400 pKa = 11.84ARR402 pKa = 11.84EE403 pKa = 3.87DD404 pKa = 3.15ASASFANSDD413 pKa = 3.29AKK415 pKa = 10.77KK416 pKa = 10.16FYY418 pKa = 11.24ADD420 pKa = 3.14
MM1 pKa = 7.92SFEE4 pKa = 4.24TSEE7 pKa = 4.11GMSRR11 pKa = 11.84PGDD14 pKa = 3.54NPNKK18 pKa = 10.32LNAKK22 pKa = 8.69PRR24 pKa = 11.84QSARR28 pKa = 11.84PKK30 pKa = 9.11TRR32 pKa = 11.84NSTAQSNQTMRR43 pKa = 11.84LGWIDD48 pKa = 4.7PLPQVDD54 pKa = 4.42TIFPLGLEE62 pKa = 4.5PNVEE66 pKa = 4.66SIPAGEE72 pKa = 4.35VEE74 pKa = 5.41LDD76 pKa = 3.53FNLPEE81 pKa = 4.38TIAKK85 pKa = 9.35PFADD89 pKa = 3.58TVTSVGDD96 pKa = 4.28RR97 pKa = 11.84IQLVDD102 pKa = 4.32DD103 pKa = 4.59DD104 pKa = 5.09KK105 pKa = 12.05EE106 pKa = 4.48NIATSIYY113 pKa = 10.37GLSFFKK119 pKa = 10.59AARR122 pKa = 11.84QLYY125 pKa = 8.14STMLDD130 pKa = 3.05HH131 pKa = 7.2EE132 pKa = 4.67KK133 pKa = 10.83AVNQPLKK140 pKa = 10.47AVYY143 pKa = 10.17YY144 pKa = 10.5DD145 pKa = 3.65EE146 pKa = 4.75TPIPAHH152 pKa = 5.97MSGALGIIGHH162 pKa = 6.41MKK164 pKa = 9.66TKK166 pKa = 10.67VGDD169 pKa = 3.93VLVKK173 pKa = 10.72DD174 pKa = 3.67AGVLFKK180 pKa = 10.72RR181 pKa = 11.84GTAAGVTKK189 pKa = 10.62FSEE192 pKa = 3.95IDD194 pKa = 3.64NDD196 pKa = 3.92KK197 pKa = 9.73TWNLDD202 pKa = 3.23CSKK205 pKa = 10.66LVWADD210 pKa = 3.45HH211 pKa = 6.66SSLSMIKK218 pKa = 10.15RR219 pKa = 11.84LASEE223 pKa = 5.13KK224 pKa = 10.18ISQLVKK230 pKa = 9.48QRR232 pKa = 11.84YY233 pKa = 8.49RR234 pKa = 11.84VTDD237 pKa = 3.71AQGHH241 pKa = 5.21VYY243 pKa = 9.78SVSMPQLTDD252 pKa = 3.15QALPDD257 pKa = 4.25YY258 pKa = 9.93YY259 pKa = 11.35DD260 pKa = 4.84SIPDD264 pKa = 3.43VAPNSDD270 pKa = 3.4QLRR273 pKa = 11.84VLTAALQMSLAQFRR287 pKa = 11.84NDD289 pKa = 3.84EE290 pKa = 4.34LPHH293 pKa = 8.28DD294 pKa = 4.55EE295 pKa = 5.32DD296 pKa = 6.08RR297 pKa = 11.84SDD299 pKa = 5.4LLTTLDD305 pKa = 4.31LLYY308 pKa = 10.93ADD310 pKa = 4.42GAYY313 pKa = 9.85EE314 pKa = 4.09ISALRR319 pKa = 11.84DD320 pKa = 3.37QFEE323 pKa = 4.79LLMARR328 pKa = 11.84YY329 pKa = 6.15TTDD332 pKa = 3.63FKK334 pKa = 11.18WRR336 pKa = 11.84VEE338 pKa = 4.52SIFKK342 pKa = 10.43VGPPPAGTTGYY353 pKa = 10.34GAQTVSSTGNTARR366 pKa = 11.84WQFPLSDD373 pKa = 3.42ADD375 pKa = 3.68INIGYY380 pKa = 9.29LFSPSKK386 pKa = 10.74SFSLFPKK393 pKa = 9.24MVGYY397 pKa = 9.88SKK399 pKa = 10.51RR400 pKa = 11.84ARR402 pKa = 11.84EE403 pKa = 3.87DD404 pKa = 3.15ASASFANSDD413 pKa = 3.29AKK415 pKa = 10.77KK416 pKa = 10.16FYY418 pKa = 11.24ADD420 pKa = 3.14
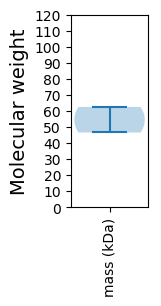
Molecular weight: 46.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4G3H2|Q4G3H2_9VIRU Putative dsRNA-dependent RNA polymerase OS=Penicillium stoloniferum virus F OX=296210 PE=4 SV=1
MM1 pKa = 7.28EE2 pKa = 4.67TTTPDD7 pKa = 3.87LPFDD11 pKa = 3.69LHH13 pKa = 5.51TRR15 pKa = 11.84EE16 pKa = 4.38AYY18 pKa = 10.06DD19 pKa = 3.12YY20 pKa = 9.61ATFHH24 pKa = 6.42RR25 pKa = 11.84TLLHH29 pKa = 6.37KK30 pKa = 10.57PGLSRR35 pKa = 11.84IKK37 pKa = 10.05EE38 pKa = 3.96DD39 pKa = 2.66RR40 pKa = 11.84WVYY43 pKa = 10.45KK44 pKa = 10.76YY45 pKa = 10.74NVEE48 pKa = 4.02QTRR51 pKa = 11.84MNTDD55 pKa = 3.09PFVRR59 pKa = 11.84KK60 pKa = 9.73SMKK63 pKa = 10.23LWDD66 pKa = 3.25EE67 pKa = 4.16HH68 pKa = 7.26AYY70 pKa = 10.66HH71 pKa = 7.51DD72 pKa = 4.1MYY74 pKa = 11.6GFTKK78 pKa = 10.43KK79 pKa = 10.42ARR81 pKa = 11.84LSNGLDD87 pKa = 3.14AFQGFAKK94 pKa = 9.82PQKK97 pKa = 9.4QRR99 pKa = 11.84SSMSPEE105 pKa = 3.53MASCYY110 pKa = 10.16EE111 pKa = 4.0KK112 pKa = 10.83ALEE115 pKa = 4.09EE116 pKa = 4.07ARR118 pKa = 11.84HH119 pKa = 5.35VFTPHH124 pKa = 5.58EE125 pKa = 4.15RR126 pKa = 11.84LTRR129 pKa = 11.84LSVPNVCDD137 pKa = 3.48STNLDD142 pKa = 3.46SAAGFSFPGKK152 pKa = 9.79KK153 pKa = 9.67KK154 pKa = 10.54SEE156 pKa = 4.24VVEE159 pKa = 4.14EE160 pKa = 4.38AFDD163 pKa = 3.62VASYY167 pKa = 10.04IAHH170 pKa = 6.29FVASDD175 pKa = 3.41RR176 pKa = 11.84KK177 pKa = 10.6VFIPPAKK184 pKa = 9.92LALRR188 pKa = 11.84GHH190 pKa = 7.07LSEE193 pKa = 4.81IDD195 pKa = 3.61EE196 pKa = 4.73LKK198 pKa = 10.07TRR200 pKa = 11.84AVWVFPFEE208 pKa = 4.18ISILEE213 pKa = 4.32GKK215 pKa = 8.26WALPYY220 pKa = 11.0YY221 pKa = 10.45KK222 pKa = 10.13FLEE225 pKa = 4.21QNVPEE230 pKa = 4.13VHH232 pKa = 6.32FGEE235 pKa = 5.28GAMQRR240 pKa = 11.84LAKK243 pKa = 9.95TLMTDD248 pKa = 3.39VASHH252 pKa = 5.55SEE254 pKa = 4.21CTEE257 pKa = 3.78VTLDD261 pKa = 3.13WSGFDD266 pKa = 3.28TSVSNWLIDD275 pKa = 3.69DD276 pKa = 4.65AFDD279 pKa = 4.2IMFDD283 pKa = 4.01SFDD286 pKa = 3.72EE287 pKa = 4.54TQVEE291 pKa = 4.08HH292 pKa = 7.42DD293 pKa = 4.06GNFVLGGDD301 pKa = 3.73HH302 pKa = 6.11MAKK305 pKa = 10.38KK306 pKa = 10.21NEE308 pKa = 4.05KK309 pKa = 9.49VKK311 pKa = 10.91KK312 pKa = 9.78FLKK315 pKa = 10.04TYY317 pKa = 9.81FKK319 pKa = 9.47KK320 pKa = 9.3TKK322 pKa = 10.15IMLPDD327 pKa = 3.89GSLYY331 pKa = 10.88KK332 pKa = 10.1KK333 pKa = 10.32FHH335 pKa = 7.06GIPSGSFFTQIIGSIVNYY353 pKa = 10.1LAVKK357 pKa = 9.34TLDD360 pKa = 5.25NYY362 pKa = 10.4FSWNARR368 pKa = 11.84RR369 pKa = 11.84FRR371 pKa = 11.84VLGDD375 pKa = 3.57DD376 pKa = 3.86SSFLIPFGRR385 pKa = 11.84SKK387 pKa = 11.29VDD389 pKa = 3.23GVEE392 pKa = 3.88ISEE395 pKa = 4.55KK396 pKa = 10.09AWEE399 pKa = 4.38TFGFTLKK406 pKa = 10.51LKK408 pKa = 10.41KK409 pKa = 10.41LRR411 pKa = 11.84IANKK415 pKa = 8.47QQDD418 pKa = 4.01RR419 pKa = 11.84KK420 pKa = 10.49FLGYY424 pKa = 9.39QCNAFRR430 pKa = 11.84YY431 pKa = 8.17EE432 pKa = 3.9RR433 pKa = 11.84STTEE437 pKa = 3.64WLSMVLYY444 pKa = 9.55PEE446 pKa = 4.73RR447 pKa = 11.84DD448 pKa = 3.55VEE450 pKa = 4.37FLEE453 pKa = 4.68QSASRR458 pKa = 11.84VFAFYY463 pKa = 11.32LLGGCNDD470 pKa = 3.02VTYY473 pKa = 11.2CEE475 pKa = 4.74FFHH478 pKa = 7.67DD479 pKa = 3.87YY480 pKa = 10.58LGRR483 pKa = 11.84YY484 pKa = 7.79PYY486 pKa = 9.95IYY488 pKa = 10.54GKK490 pKa = 9.55EE491 pKa = 3.97LPLTRR496 pKa = 11.84GLKK499 pKa = 10.21RR500 pKa = 11.84LFKK503 pKa = 10.61FVFRR507 pKa = 11.84LTIDD511 pKa = 3.1KK512 pKa = 10.68LAFPDD517 pKa = 4.38LSRR520 pKa = 11.84FDD522 pKa = 3.93PLKK525 pKa = 11.12VPFSLSLGDD534 pKa = 3.56KK535 pKa = 10.15PFWW538 pKa = 4.63
MM1 pKa = 7.28EE2 pKa = 4.67TTTPDD7 pKa = 3.87LPFDD11 pKa = 3.69LHH13 pKa = 5.51TRR15 pKa = 11.84EE16 pKa = 4.38AYY18 pKa = 10.06DD19 pKa = 3.12YY20 pKa = 9.61ATFHH24 pKa = 6.42RR25 pKa = 11.84TLLHH29 pKa = 6.37KK30 pKa = 10.57PGLSRR35 pKa = 11.84IKK37 pKa = 10.05EE38 pKa = 3.96DD39 pKa = 2.66RR40 pKa = 11.84WVYY43 pKa = 10.45KK44 pKa = 10.76YY45 pKa = 10.74NVEE48 pKa = 4.02QTRR51 pKa = 11.84MNTDD55 pKa = 3.09PFVRR59 pKa = 11.84KK60 pKa = 9.73SMKK63 pKa = 10.23LWDD66 pKa = 3.25EE67 pKa = 4.16HH68 pKa = 7.26AYY70 pKa = 10.66HH71 pKa = 7.51DD72 pKa = 4.1MYY74 pKa = 11.6GFTKK78 pKa = 10.43KK79 pKa = 10.42ARR81 pKa = 11.84LSNGLDD87 pKa = 3.14AFQGFAKK94 pKa = 9.82PQKK97 pKa = 9.4QRR99 pKa = 11.84SSMSPEE105 pKa = 3.53MASCYY110 pKa = 10.16EE111 pKa = 4.0KK112 pKa = 10.83ALEE115 pKa = 4.09EE116 pKa = 4.07ARR118 pKa = 11.84HH119 pKa = 5.35VFTPHH124 pKa = 5.58EE125 pKa = 4.15RR126 pKa = 11.84LTRR129 pKa = 11.84LSVPNVCDD137 pKa = 3.48STNLDD142 pKa = 3.46SAAGFSFPGKK152 pKa = 9.79KK153 pKa = 9.67KK154 pKa = 10.54SEE156 pKa = 4.24VVEE159 pKa = 4.14EE160 pKa = 4.38AFDD163 pKa = 3.62VASYY167 pKa = 10.04IAHH170 pKa = 6.29FVASDD175 pKa = 3.41RR176 pKa = 11.84KK177 pKa = 10.6VFIPPAKK184 pKa = 9.92LALRR188 pKa = 11.84GHH190 pKa = 7.07LSEE193 pKa = 4.81IDD195 pKa = 3.61EE196 pKa = 4.73LKK198 pKa = 10.07TRR200 pKa = 11.84AVWVFPFEE208 pKa = 4.18ISILEE213 pKa = 4.32GKK215 pKa = 8.26WALPYY220 pKa = 11.0YY221 pKa = 10.45KK222 pKa = 10.13FLEE225 pKa = 4.21QNVPEE230 pKa = 4.13VHH232 pKa = 6.32FGEE235 pKa = 5.28GAMQRR240 pKa = 11.84LAKK243 pKa = 9.95TLMTDD248 pKa = 3.39VASHH252 pKa = 5.55SEE254 pKa = 4.21CTEE257 pKa = 3.78VTLDD261 pKa = 3.13WSGFDD266 pKa = 3.28TSVSNWLIDD275 pKa = 3.69DD276 pKa = 4.65AFDD279 pKa = 4.2IMFDD283 pKa = 4.01SFDD286 pKa = 3.72EE287 pKa = 4.54TQVEE291 pKa = 4.08HH292 pKa = 7.42DD293 pKa = 4.06GNFVLGGDD301 pKa = 3.73HH302 pKa = 6.11MAKK305 pKa = 10.38KK306 pKa = 10.21NEE308 pKa = 4.05KK309 pKa = 9.49VKK311 pKa = 10.91KK312 pKa = 9.78FLKK315 pKa = 10.04TYY317 pKa = 9.81FKK319 pKa = 9.47KK320 pKa = 9.3TKK322 pKa = 10.15IMLPDD327 pKa = 3.89GSLYY331 pKa = 10.88KK332 pKa = 10.1KK333 pKa = 10.32FHH335 pKa = 7.06GIPSGSFFTQIIGSIVNYY353 pKa = 10.1LAVKK357 pKa = 9.34TLDD360 pKa = 5.25NYY362 pKa = 10.4FSWNARR368 pKa = 11.84RR369 pKa = 11.84FRR371 pKa = 11.84VLGDD375 pKa = 3.57DD376 pKa = 3.86SSFLIPFGRR385 pKa = 11.84SKK387 pKa = 11.29VDD389 pKa = 3.23GVEE392 pKa = 3.88ISEE395 pKa = 4.55KK396 pKa = 10.09AWEE399 pKa = 4.38TFGFTLKK406 pKa = 10.51LKK408 pKa = 10.41KK409 pKa = 10.41LRR411 pKa = 11.84IANKK415 pKa = 8.47QQDD418 pKa = 4.01RR419 pKa = 11.84KK420 pKa = 10.49FLGYY424 pKa = 9.39QCNAFRR430 pKa = 11.84YY431 pKa = 8.17EE432 pKa = 3.9RR433 pKa = 11.84STTEE437 pKa = 3.64WLSMVLYY444 pKa = 9.55PEE446 pKa = 4.73RR447 pKa = 11.84DD448 pKa = 3.55VEE450 pKa = 4.37FLEE453 pKa = 4.68QSASRR458 pKa = 11.84VFAFYY463 pKa = 11.32LLGGCNDD470 pKa = 3.02VTYY473 pKa = 11.2CEE475 pKa = 4.74FFHH478 pKa = 7.67DD479 pKa = 3.87YY480 pKa = 10.58LGRR483 pKa = 11.84YY484 pKa = 7.79PYY486 pKa = 9.95IYY488 pKa = 10.54GKK490 pKa = 9.55EE491 pKa = 3.97LPLTRR496 pKa = 11.84GLKK499 pKa = 10.21RR500 pKa = 11.84LFKK503 pKa = 10.61FVFRR507 pKa = 11.84LTIDD511 pKa = 3.1KK512 pKa = 10.68LAFPDD517 pKa = 4.38LSRR520 pKa = 11.84FDD522 pKa = 3.93PLKK525 pKa = 11.12VPFSLSLGDD534 pKa = 3.56KK535 pKa = 10.15PFWW538 pKa = 4.63
Molecular weight: 62.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
958 |
420 |
538 |
479.0 |
54.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.307 ± 1.152 | 0.731 ± 0.326 |
7.516 ± 0.699 | 5.532 ± 0.825 |
6.681 ± 1.585 | 5.428 ± 0.032 |
2.192 ± 0.505 | 3.758 ± 0.349 |
7.203 ± 0.827 | 9.29 ± 0.161 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.401 ± 0.144 | 3.236 ± 0.38 |
5.115 ± 0.554 | 3.132 ± 0.764 |
5.219 ± 0.303 | 7.933 ± 0.738 |
5.95 ± 0.317 | 5.846 ± 0.087 |
1.566 ± 0.248 | 3.967 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
