
Night heron coronavirus HKU19
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Deltacoronavirus; Herdecovirus
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|H9BR18|H9BR18_9NIDO Envelope protein OS=Night heron coronavirus HKU19 OX=1159904 GN=E PE=4 SV=1
MM1 pKa = 6.73QCVVLVLTLISIVTSRR17 pKa = 11.84PNSFADD23 pKa = 3.51RR24 pKa = 11.84VFDD27 pKa = 4.78ALTFPHH33 pKa = 7.18ASNYY37 pKa = 9.78LHH39 pKa = 6.9VGDD42 pKa = 4.81KK43 pKa = 10.41TPSRR47 pKa = 11.84PQLLQARR54 pKa = 11.84NQGNYY59 pKa = 9.86SAICPEE65 pKa = 3.77NGYY68 pKa = 8.28ITSTSYY74 pKa = 11.31DD75 pKa = 3.36LSKK78 pKa = 10.42IYY80 pKa = 10.82YY81 pKa = 8.25LTDD84 pKa = 2.93GDD86 pKa = 4.24YY87 pKa = 11.13PIDD90 pKa = 3.72GVYY93 pKa = 10.68KK94 pKa = 10.4SLQPLKK100 pKa = 9.37VTCVPEE106 pKa = 3.47WHH108 pKa = 6.96GNSNFNDD115 pKa = 3.54TTGWKK120 pKa = 9.78HH121 pKa = 5.67YY122 pKa = 10.19FDD124 pKa = 5.57GRR126 pKa = 11.84IKK128 pKa = 10.67QNPNTIWCPCSQSGPGGAQHH148 pKa = 6.51NAAGNSTEE156 pKa = 3.97YY157 pKa = 10.42IRR159 pKa = 11.84FHH161 pKa = 8.1SNITTSVSNLLRR173 pKa = 11.84LYY175 pKa = 10.86AVDD178 pKa = 3.05NQYY181 pKa = 11.55YY182 pKa = 9.72YY183 pKa = 10.6FGCTPTPTPLTFNLTSEE200 pKa = 4.54NITLFTAEE208 pKa = 4.3EE209 pKa = 4.07QVHH212 pKa = 5.27YY213 pKa = 10.34CYY215 pKa = 11.31ANINGTVSYY224 pKa = 10.54IGVLPPKK231 pKa = 9.51ITEE234 pKa = 4.24LTVGRR239 pKa = 11.84YY240 pKa = 9.59GDD242 pKa = 3.34IFVNGFLYY250 pKa = 9.95FKK252 pKa = 9.93IPNVIEE258 pKa = 4.06YY259 pKa = 9.73VQLSHH264 pKa = 7.46TIPHH268 pKa = 6.27NKK270 pKa = 8.81QFYY273 pKa = 8.24TVFYY277 pKa = 11.23ANMTQVLLNISMASINRR294 pKa = 11.84LLYY297 pKa = 10.13CDD299 pKa = 3.92KK300 pKa = 11.02DD301 pKa = 4.13SYY303 pKa = 11.86SSIACAVNQFEE314 pKa = 4.66PANGFYY320 pKa = 9.56STSAIEE326 pKa = 4.26KK327 pKa = 9.06ISRR330 pKa = 11.84KK331 pKa = 7.79FTFVTMPTVQNHH343 pKa = 5.75SYY345 pKa = 11.16YY346 pKa = 10.71SINLTIGGCGHH357 pKa = 6.92GEE359 pKa = 3.96YY360 pKa = 9.9PHH362 pKa = 7.01LSNKK366 pKa = 7.61TGCYY370 pKa = 8.57RR371 pKa = 11.84TDD373 pKa = 3.09ASNINAKK380 pKa = 10.34QITFVINTYY389 pKa = 7.58THH391 pKa = 7.18DD392 pKa = 3.78NWIQWAHH399 pKa = 6.52KK400 pKa = 9.87PGNCPWALNKK410 pKa = 9.97INNYY414 pKa = 7.36NTAGTLQVVPEE425 pKa = 4.13NQATCCTDD433 pKa = 3.35NQASWLYY440 pKa = 9.08LASWTSVNVKK450 pKa = 9.8VCFNYY455 pKa = 10.16QPGTTISIQPQQTGVATDD473 pKa = 3.25ISVIYY478 pKa = 10.42EE479 pKa = 4.17NEE481 pKa = 4.01CVDD484 pKa = 3.41YY485 pKa = 11.21NIYY488 pKa = 10.79GKK490 pKa = 8.52TGTGIIQSTNVTLLAGRR507 pKa = 11.84TYY509 pKa = 10.83TSASGQLLAFKK520 pKa = 10.28YY521 pKa = 10.57LSNQTIYY528 pKa = 11.19SVTPCDD534 pKa = 3.94FSNQVAVYY542 pKa = 8.52NKK544 pKa = 10.37SVIAAILPQNKK555 pKa = 9.01TIFGLTNIQEE565 pKa = 4.46TPNFYY570 pKa = 10.23IANNAHH576 pKa = 4.48QQQRR580 pKa = 11.84FAMYY584 pKa = 9.19MEE586 pKa = 5.27PLNSQQPDD594 pKa = 3.76CTPVLTYY601 pKa = 10.8AQIGICADD609 pKa = 3.51GQFVQVQPEE618 pKa = 4.1KK619 pKa = 9.64SQPMSTTPIVAVNITIPKK637 pKa = 7.65TFNISVQTEE646 pKa = 4.14YY647 pKa = 10.83IQISTDD653 pKa = 3.61NIVIDD658 pKa = 4.05CQRR661 pKa = 11.84YY662 pKa = 7.95VCNGNPRR669 pKa = 11.84CLMLLSQYY677 pKa = 10.88QSACSTIEE685 pKa = 3.73QALHH689 pKa = 4.66QKK691 pKa = 10.28ARR693 pKa = 11.84LEE695 pKa = 3.98SLEE698 pKa = 4.29LSTMLAYY705 pKa = 10.54SPNTLQLANVSNFQSNNMGFNLTNLLPQNNSPQKK739 pKa = 10.12RR740 pKa = 11.84SVIEE744 pKa = 4.18DD745 pKa = 3.59LLFSKK750 pKa = 10.81VVTNGLGTVDD760 pKa = 3.92VDD762 pKa = 4.09YY763 pKa = 11.28KK764 pKa = 11.13KK765 pKa = 9.82CTKK768 pKa = 10.11GLSIADD774 pKa = 4.3LPCAQYY780 pKa = 11.51YY781 pKa = 9.18NGIMVLPGVADD792 pKa = 3.69SGLLAAYY799 pKa = 7.24TASLTGGMVFGGLTSAAAIPFATAVQARR827 pKa = 11.84LNYY830 pKa = 9.72VALQTDD836 pKa = 3.94VLQRR840 pKa = 11.84NQQILANAFNQAMGNITLAFKK861 pKa = 10.39DD862 pKa = 3.66VKK864 pKa = 10.53EE865 pKa = 4.95AIATTADD872 pKa = 4.33AIRR875 pKa = 11.84VVAGALDD882 pKa = 4.87KK883 pKa = 10.83IQQVVNSQGQALSKK897 pKa = 10.15LTGEE901 pKa = 4.23LQRR904 pKa = 11.84NFQAISASIEE914 pKa = 4.2DD915 pKa = 3.63IYY917 pKa = 11.83NRR919 pKa = 11.84LNDD922 pKa = 3.64IEE924 pKa = 6.43ADD926 pKa = 3.62AQVDD930 pKa = 3.69RR931 pKa = 11.84LITGRR936 pKa = 11.84LAALNAFLTQTLTQANEE953 pKa = 3.69VKK955 pKa = 10.47AARR958 pKa = 11.84EE959 pKa = 3.65LALQKK964 pKa = 10.48INEE967 pKa = 4.38CVKK970 pKa = 10.66DD971 pKa = 3.42QSKK974 pKa = 10.82RR975 pKa = 11.84YY976 pKa = 7.99GFCGNGYY983 pKa = 9.62HH984 pKa = 7.05LFSIANAAPKK994 pKa = 10.69GFIFFHH1000 pKa = 6.07TVLQPEE1006 pKa = 4.43TTIEE1010 pKa = 3.8IQAIAGFCVSDD1021 pKa = 3.32RR1022 pKa = 11.84QTYY1025 pKa = 10.01NYY1027 pKa = 9.94YY1028 pKa = 10.08SSNMEE1033 pKa = 3.58GQAYY1037 pKa = 9.18IARR1040 pKa = 11.84DD1041 pKa = 3.53TTQTIFLHH1049 pKa = 6.37EE1050 pKa = 4.87NGTYY1054 pKa = 9.71MITPRR1059 pKa = 11.84KK1060 pKa = 8.96QYY1062 pKa = 9.8QPRR1065 pKa = 11.84TLAQADD1071 pKa = 3.8VVKK1074 pKa = 10.79ISTCDD1079 pKa = 3.13VTYY1082 pKa = 11.46VNLTSIEE1089 pKa = 4.1FEE1091 pKa = 4.05QLIPEE1096 pKa = 4.2YY1097 pKa = 11.05VDD1099 pKa = 3.03INSTVEE1105 pKa = 4.4GILNSTLPGKK1115 pKa = 10.37IPDD1118 pKa = 4.29LNIGHH1123 pKa = 6.4YY1124 pKa = 11.07NNTILNLTTEE1134 pKa = 4.25INDD1137 pKa = 3.86LQSKK1141 pKa = 10.57AEE1143 pKa = 4.12NLSMIAYY1150 pKa = 9.69QLEE1153 pKa = 4.4EE1154 pKa = 4.24YY1155 pKa = 10.04IKK1157 pKa = 10.76NINNTLVDD1165 pKa = 4.54LEE1167 pKa = 4.17WLNRR1171 pKa = 11.84VEE1173 pKa = 5.99TYY1175 pKa = 10.68LKK1177 pKa = 9.3WPWYY1181 pKa = 8.42VWLAIALAFTGFVTILITIFLCTGCCGGCFGCCGGCFGLFSKK1223 pKa = 10.66KK1224 pKa = 9.19IDD1226 pKa = 3.62PMRR1229 pKa = 11.84QYY1231 pKa = 10.23MNRR1234 pKa = 11.84YY1235 pKa = 5.86EE1236 pKa = 4.29TPTSKK1241 pKa = 10.83SDD1243 pKa = 3.39DD1244 pKa = 4.87AIPIIYY1250 pKa = 9.43KK1251 pKa = 10.59KK1252 pKa = 10.54NWW1254 pKa = 3.01
MM1 pKa = 6.73QCVVLVLTLISIVTSRR17 pKa = 11.84PNSFADD23 pKa = 3.51RR24 pKa = 11.84VFDD27 pKa = 4.78ALTFPHH33 pKa = 7.18ASNYY37 pKa = 9.78LHH39 pKa = 6.9VGDD42 pKa = 4.81KK43 pKa = 10.41TPSRR47 pKa = 11.84PQLLQARR54 pKa = 11.84NQGNYY59 pKa = 9.86SAICPEE65 pKa = 3.77NGYY68 pKa = 8.28ITSTSYY74 pKa = 11.31DD75 pKa = 3.36LSKK78 pKa = 10.42IYY80 pKa = 10.82YY81 pKa = 8.25LTDD84 pKa = 2.93GDD86 pKa = 4.24YY87 pKa = 11.13PIDD90 pKa = 3.72GVYY93 pKa = 10.68KK94 pKa = 10.4SLQPLKK100 pKa = 9.37VTCVPEE106 pKa = 3.47WHH108 pKa = 6.96GNSNFNDD115 pKa = 3.54TTGWKK120 pKa = 9.78HH121 pKa = 5.67YY122 pKa = 10.19FDD124 pKa = 5.57GRR126 pKa = 11.84IKK128 pKa = 10.67QNPNTIWCPCSQSGPGGAQHH148 pKa = 6.51NAAGNSTEE156 pKa = 3.97YY157 pKa = 10.42IRR159 pKa = 11.84FHH161 pKa = 8.1SNITTSVSNLLRR173 pKa = 11.84LYY175 pKa = 10.86AVDD178 pKa = 3.05NQYY181 pKa = 11.55YY182 pKa = 9.72YY183 pKa = 10.6FGCTPTPTPLTFNLTSEE200 pKa = 4.54NITLFTAEE208 pKa = 4.3EE209 pKa = 4.07QVHH212 pKa = 5.27YY213 pKa = 10.34CYY215 pKa = 11.31ANINGTVSYY224 pKa = 10.54IGVLPPKK231 pKa = 9.51ITEE234 pKa = 4.24LTVGRR239 pKa = 11.84YY240 pKa = 9.59GDD242 pKa = 3.34IFVNGFLYY250 pKa = 9.95FKK252 pKa = 9.93IPNVIEE258 pKa = 4.06YY259 pKa = 9.73VQLSHH264 pKa = 7.46TIPHH268 pKa = 6.27NKK270 pKa = 8.81QFYY273 pKa = 8.24TVFYY277 pKa = 11.23ANMTQVLLNISMASINRR294 pKa = 11.84LLYY297 pKa = 10.13CDD299 pKa = 3.92KK300 pKa = 11.02DD301 pKa = 4.13SYY303 pKa = 11.86SSIACAVNQFEE314 pKa = 4.66PANGFYY320 pKa = 9.56STSAIEE326 pKa = 4.26KK327 pKa = 9.06ISRR330 pKa = 11.84KK331 pKa = 7.79FTFVTMPTVQNHH343 pKa = 5.75SYY345 pKa = 11.16YY346 pKa = 10.71SINLTIGGCGHH357 pKa = 6.92GEE359 pKa = 3.96YY360 pKa = 9.9PHH362 pKa = 7.01LSNKK366 pKa = 7.61TGCYY370 pKa = 8.57RR371 pKa = 11.84TDD373 pKa = 3.09ASNINAKK380 pKa = 10.34QITFVINTYY389 pKa = 7.58THH391 pKa = 7.18DD392 pKa = 3.78NWIQWAHH399 pKa = 6.52KK400 pKa = 9.87PGNCPWALNKK410 pKa = 9.97INNYY414 pKa = 7.36NTAGTLQVVPEE425 pKa = 4.13NQATCCTDD433 pKa = 3.35NQASWLYY440 pKa = 9.08LASWTSVNVKK450 pKa = 9.8VCFNYY455 pKa = 10.16QPGTTISIQPQQTGVATDD473 pKa = 3.25ISVIYY478 pKa = 10.42EE479 pKa = 4.17NEE481 pKa = 4.01CVDD484 pKa = 3.41YY485 pKa = 11.21NIYY488 pKa = 10.79GKK490 pKa = 8.52TGTGIIQSTNVTLLAGRR507 pKa = 11.84TYY509 pKa = 10.83TSASGQLLAFKK520 pKa = 10.28YY521 pKa = 10.57LSNQTIYY528 pKa = 11.19SVTPCDD534 pKa = 3.94FSNQVAVYY542 pKa = 8.52NKK544 pKa = 10.37SVIAAILPQNKK555 pKa = 9.01TIFGLTNIQEE565 pKa = 4.46TPNFYY570 pKa = 10.23IANNAHH576 pKa = 4.48QQQRR580 pKa = 11.84FAMYY584 pKa = 9.19MEE586 pKa = 5.27PLNSQQPDD594 pKa = 3.76CTPVLTYY601 pKa = 10.8AQIGICADD609 pKa = 3.51GQFVQVQPEE618 pKa = 4.1KK619 pKa = 9.64SQPMSTTPIVAVNITIPKK637 pKa = 7.65TFNISVQTEE646 pKa = 4.14YY647 pKa = 10.83IQISTDD653 pKa = 3.61NIVIDD658 pKa = 4.05CQRR661 pKa = 11.84YY662 pKa = 7.95VCNGNPRR669 pKa = 11.84CLMLLSQYY677 pKa = 10.88QSACSTIEE685 pKa = 3.73QALHH689 pKa = 4.66QKK691 pKa = 10.28ARR693 pKa = 11.84LEE695 pKa = 3.98SLEE698 pKa = 4.29LSTMLAYY705 pKa = 10.54SPNTLQLANVSNFQSNNMGFNLTNLLPQNNSPQKK739 pKa = 10.12RR740 pKa = 11.84SVIEE744 pKa = 4.18DD745 pKa = 3.59LLFSKK750 pKa = 10.81VVTNGLGTVDD760 pKa = 3.92VDD762 pKa = 4.09YY763 pKa = 11.28KK764 pKa = 11.13KK765 pKa = 9.82CTKK768 pKa = 10.11GLSIADD774 pKa = 4.3LPCAQYY780 pKa = 11.51YY781 pKa = 9.18NGIMVLPGVADD792 pKa = 3.69SGLLAAYY799 pKa = 7.24TASLTGGMVFGGLTSAAAIPFATAVQARR827 pKa = 11.84LNYY830 pKa = 9.72VALQTDD836 pKa = 3.94VLQRR840 pKa = 11.84NQQILANAFNQAMGNITLAFKK861 pKa = 10.39DD862 pKa = 3.66VKK864 pKa = 10.53EE865 pKa = 4.95AIATTADD872 pKa = 4.33AIRR875 pKa = 11.84VVAGALDD882 pKa = 4.87KK883 pKa = 10.83IQQVVNSQGQALSKK897 pKa = 10.15LTGEE901 pKa = 4.23LQRR904 pKa = 11.84NFQAISASIEE914 pKa = 4.2DD915 pKa = 3.63IYY917 pKa = 11.83NRR919 pKa = 11.84LNDD922 pKa = 3.64IEE924 pKa = 6.43ADD926 pKa = 3.62AQVDD930 pKa = 3.69RR931 pKa = 11.84LITGRR936 pKa = 11.84LAALNAFLTQTLTQANEE953 pKa = 3.69VKK955 pKa = 10.47AARR958 pKa = 11.84EE959 pKa = 3.65LALQKK964 pKa = 10.48INEE967 pKa = 4.38CVKK970 pKa = 10.66DD971 pKa = 3.42QSKK974 pKa = 10.82RR975 pKa = 11.84YY976 pKa = 7.99GFCGNGYY983 pKa = 9.62HH984 pKa = 7.05LFSIANAAPKK994 pKa = 10.69GFIFFHH1000 pKa = 6.07TVLQPEE1006 pKa = 4.43TTIEE1010 pKa = 3.8IQAIAGFCVSDD1021 pKa = 3.32RR1022 pKa = 11.84QTYY1025 pKa = 10.01NYY1027 pKa = 9.94YY1028 pKa = 10.08SSNMEE1033 pKa = 3.58GQAYY1037 pKa = 9.18IARR1040 pKa = 11.84DD1041 pKa = 3.53TTQTIFLHH1049 pKa = 6.37EE1050 pKa = 4.87NGTYY1054 pKa = 9.71MITPRR1059 pKa = 11.84KK1060 pKa = 8.96QYY1062 pKa = 9.8QPRR1065 pKa = 11.84TLAQADD1071 pKa = 3.8VVKK1074 pKa = 10.79ISTCDD1079 pKa = 3.13VTYY1082 pKa = 11.46VNLTSIEE1089 pKa = 4.1FEE1091 pKa = 4.05QLIPEE1096 pKa = 4.2YY1097 pKa = 11.05VDD1099 pKa = 3.03INSTVEE1105 pKa = 4.4GILNSTLPGKK1115 pKa = 10.37IPDD1118 pKa = 4.29LNIGHH1123 pKa = 6.4YY1124 pKa = 11.07NNTILNLTTEE1134 pKa = 4.25INDD1137 pKa = 3.86LQSKK1141 pKa = 10.57AEE1143 pKa = 4.12NLSMIAYY1150 pKa = 9.69QLEE1153 pKa = 4.4EE1154 pKa = 4.24YY1155 pKa = 10.04IKK1157 pKa = 10.76NINNTLVDD1165 pKa = 4.54LEE1167 pKa = 4.17WLNRR1171 pKa = 11.84VEE1173 pKa = 5.99TYY1175 pKa = 10.68LKK1177 pKa = 9.3WPWYY1181 pKa = 8.42VWLAIALAFTGFVTILITIFLCTGCCGGCFGCCGGCFGLFSKK1223 pKa = 10.66KK1224 pKa = 9.19IDD1226 pKa = 3.62PMRR1229 pKa = 11.84QYY1231 pKa = 10.23MNRR1234 pKa = 11.84YY1235 pKa = 5.86EE1236 pKa = 4.29TPTSKK1241 pKa = 10.83SDD1243 pKa = 3.39DD1244 pKa = 4.87AIPIIYY1250 pKa = 9.43KK1251 pKa = 10.59KK1252 pKa = 10.54NWW1254 pKa = 3.01
Molecular weight: 139.51 kDa
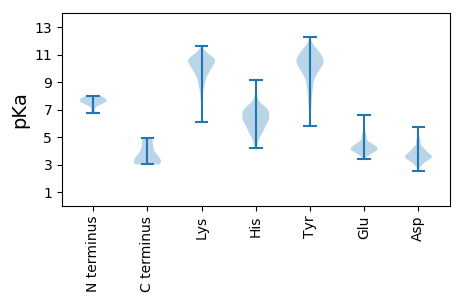
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9BR22|H9BR22_9NIDO NS7a protein OS=Night heron coronavirus HKU19 OX=1159904 GN=NS7a PE=4 SV=1
MM1 pKa = 7.58AVPRR5 pKa = 11.84VPRR8 pKa = 11.84ADD10 pKa = 3.59ASWFQPIKK18 pKa = 10.52AQNKK22 pKa = 9.59KK23 pKa = 8.02ITAPSFKK30 pKa = 10.65KK31 pKa = 10.62NGVPVNASLQAADD44 pKa = 3.35QHH46 pKa = 6.62GYY48 pKa = 6.39WLRR51 pKa = 11.84YY52 pKa = 9.34NRR54 pKa = 11.84TKK56 pKa = 10.79SGGGSIPAAFSFYY69 pKa = 9.81YY70 pKa = 10.29TGTGPYY76 pKa = 8.4ATLKK80 pKa = 10.76FGEE83 pKa = 4.58TPSDD87 pKa = 3.59RR88 pKa = 11.84ITWVKK93 pKa = 10.46GPSAVTTNKK102 pKa = 10.04PKK104 pKa = 10.09VAKK107 pKa = 9.82RR108 pKa = 11.84DD109 pKa = 3.82PDD111 pKa = 3.53KK112 pKa = 11.45HH113 pKa = 6.25PLLPLTFPPGDD124 pKa = 3.53GVNTGLRR131 pKa = 11.84VDD133 pKa = 4.14PFQNRR138 pKa = 11.84GRR140 pKa = 11.84TMDD143 pKa = 3.78RR144 pKa = 11.84GPNQRR149 pKa = 11.84SSSAPVDD156 pKa = 3.43SGNRR160 pKa = 11.84NQRR163 pKa = 11.84ARR165 pKa = 11.84SKK167 pKa = 10.38SAPSRR172 pKa = 11.84TNPKK176 pKa = 8.0THH178 pKa = 6.58QIPKK182 pKa = 8.71RR183 pKa = 11.84TLTKK187 pKa = 10.51GKK189 pKa = 8.71TISQVFGKK197 pKa = 8.61RR198 pKa = 11.84TPGGANVGSADD209 pKa = 3.67TEE211 pKa = 4.23KK212 pKa = 11.17AGFTDD217 pKa = 3.7PRR219 pKa = 11.84LMCLMRR225 pKa = 11.84HH226 pKa = 4.95TPGTQEE232 pKa = 4.33LLLAGHH238 pKa = 7.79LDD240 pKa = 3.59HH241 pKa = 7.89DD242 pKa = 4.09IQPEE246 pKa = 4.26GVYY249 pKa = 10.85LKK251 pKa = 9.3FTYY254 pKa = 7.52TTQVKK259 pKa = 9.81RR260 pKa = 11.84DD261 pKa = 3.29SAEE264 pKa = 3.74YY265 pKa = 10.68DD266 pKa = 3.37RR267 pKa = 11.84VIAALNNVVNQKK279 pKa = 10.27YY280 pKa = 10.11DD281 pKa = 3.59ADD283 pKa = 4.04NLPSQKK289 pKa = 10.13PKK291 pKa = 9.2STKK294 pKa = 9.88PKK296 pKa = 8.89PKK298 pKa = 9.77KK299 pKa = 9.22EE300 pKa = 4.16KK301 pKa = 10.4KK302 pKa = 9.48EE303 pKa = 3.79RR304 pKa = 11.84APPKK308 pKa = 9.48QQQKK312 pKa = 7.24EE313 pKa = 4.24QKK315 pKa = 9.74QEE317 pKa = 3.93PQHH320 pKa = 6.06EE321 pKa = 4.51SEE323 pKa = 4.5DD324 pKa = 3.68TKK326 pKa = 10.88KK327 pKa = 10.53IQWAEE332 pKa = 3.91DD333 pKa = 3.55DD334 pKa = 3.84TDD336 pKa = 4.1FAASFSS342 pKa = 3.85
MM1 pKa = 7.58AVPRR5 pKa = 11.84VPRR8 pKa = 11.84ADD10 pKa = 3.59ASWFQPIKK18 pKa = 10.52AQNKK22 pKa = 9.59KK23 pKa = 8.02ITAPSFKK30 pKa = 10.65KK31 pKa = 10.62NGVPVNASLQAADD44 pKa = 3.35QHH46 pKa = 6.62GYY48 pKa = 6.39WLRR51 pKa = 11.84YY52 pKa = 9.34NRR54 pKa = 11.84TKK56 pKa = 10.79SGGGSIPAAFSFYY69 pKa = 9.81YY70 pKa = 10.29TGTGPYY76 pKa = 8.4ATLKK80 pKa = 10.76FGEE83 pKa = 4.58TPSDD87 pKa = 3.59RR88 pKa = 11.84ITWVKK93 pKa = 10.46GPSAVTTNKK102 pKa = 10.04PKK104 pKa = 10.09VAKK107 pKa = 9.82RR108 pKa = 11.84DD109 pKa = 3.82PDD111 pKa = 3.53KK112 pKa = 11.45HH113 pKa = 6.25PLLPLTFPPGDD124 pKa = 3.53GVNTGLRR131 pKa = 11.84VDD133 pKa = 4.14PFQNRR138 pKa = 11.84GRR140 pKa = 11.84TMDD143 pKa = 3.78RR144 pKa = 11.84GPNQRR149 pKa = 11.84SSSAPVDD156 pKa = 3.43SGNRR160 pKa = 11.84NQRR163 pKa = 11.84ARR165 pKa = 11.84SKK167 pKa = 10.38SAPSRR172 pKa = 11.84TNPKK176 pKa = 8.0THH178 pKa = 6.58QIPKK182 pKa = 8.71RR183 pKa = 11.84TLTKK187 pKa = 10.51GKK189 pKa = 8.71TISQVFGKK197 pKa = 8.61RR198 pKa = 11.84TPGGANVGSADD209 pKa = 3.67TEE211 pKa = 4.23KK212 pKa = 11.17AGFTDD217 pKa = 3.7PRR219 pKa = 11.84LMCLMRR225 pKa = 11.84HH226 pKa = 4.95TPGTQEE232 pKa = 4.33LLLAGHH238 pKa = 7.79LDD240 pKa = 3.59HH241 pKa = 7.89DD242 pKa = 4.09IQPEE246 pKa = 4.26GVYY249 pKa = 10.85LKK251 pKa = 9.3FTYY254 pKa = 7.52TTQVKK259 pKa = 9.81RR260 pKa = 11.84DD261 pKa = 3.29SAEE264 pKa = 3.74YY265 pKa = 10.68DD266 pKa = 3.37RR267 pKa = 11.84VIAALNNVVNQKK279 pKa = 10.27YY280 pKa = 10.11DD281 pKa = 3.59ADD283 pKa = 4.04NLPSQKK289 pKa = 10.13PKK291 pKa = 9.2STKK294 pKa = 9.88PKK296 pKa = 8.89PKK298 pKa = 9.77KK299 pKa = 9.22EE300 pKa = 4.16KK301 pKa = 10.4KK302 pKa = 9.48EE303 pKa = 3.79RR304 pKa = 11.84APPKK308 pKa = 9.48QQQKK312 pKa = 7.24EE313 pKa = 4.24QKK315 pKa = 9.74QEE317 pKa = 3.93PQHH320 pKa = 6.06EE321 pKa = 4.51SEE323 pKa = 4.5DD324 pKa = 3.68TKK326 pKa = 10.88KK327 pKa = 10.53IQWAEE332 pKa = 3.91DD333 pKa = 3.55DD334 pKa = 3.84TDD336 pKa = 4.1FAASFSS342 pKa = 3.85
Molecular weight: 37.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8461 |
82 |
6280 |
1057.6 |
118.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.489 ± 0.421 | 2.565 ± 0.269 |
5.46 ± 0.632 | 3.735 ± 0.328 |
4.29 ± 0.233 | 5.035 ± 0.418 |
2.269 ± 0.256 | 6.288 ± 0.667 |
5.874 ± 0.701 | 8.982 ± 0.777 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.116 ± 0.299 | 6.217 ± 0.924 |
4.278 ± 0.679 | 4.16 ± 0.735 |
2.801 ± 0.485 | 6.56 ± 0.463 |
8.415 ± 0.263 | 8.013 ± 0.926 |
0.993 ± 0.471 | 5.46 ± 0.403 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
