
Avon-Heathcote Estuary associated circular virus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
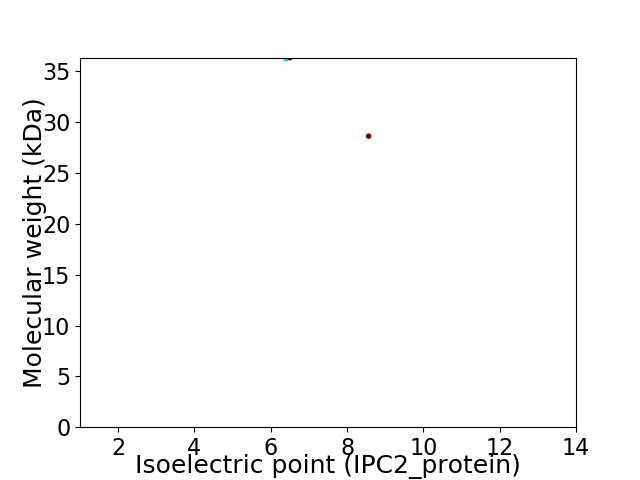
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IM93|A0A0C5IM93_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 5 OX=1618256 PE=3 SV=1
MM1 pKa = 7.85AANQKK6 pKa = 9.52FKK8 pKa = 11.13NVCFTLNNPGEE19 pKa = 4.28DD20 pKa = 4.27PIPFDD25 pKa = 3.81VEE27 pKa = 3.76KK28 pKa = 10.0MHH30 pKa = 6.15YY31 pKa = 9.75LVYY34 pKa = 10.33QRR36 pKa = 11.84EE37 pKa = 4.2VGANGTEE44 pKa = 4.49HH45 pKa = 5.71YY46 pKa = 10.19QGYY49 pKa = 9.87CEE51 pKa = 5.9LINQTRR57 pKa = 11.84FNAVKK62 pKa = 10.14ALLGGGSVHH71 pKa = 6.44IEE73 pKa = 3.77RR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 10.69GSAQEE81 pKa = 3.41AAAYY85 pKa = 8.51CKK87 pKa = 10.34KK88 pKa = 10.66DD89 pKa = 3.45DD90 pKa = 3.77TRR92 pKa = 11.84APGASIQEE100 pKa = 4.29FGEE103 pKa = 4.09MHH105 pKa = 6.99ACTPGKK111 pKa = 10.64RR112 pKa = 11.84NDD114 pKa = 3.42LKK116 pKa = 11.47AFVDD120 pKa = 4.34DD121 pKa = 3.97VRR123 pKa = 11.84SGTKK127 pKa = 9.72RR128 pKa = 11.84KK129 pKa = 9.36RR130 pKa = 11.84DD131 pKa = 3.62LVEE134 pKa = 3.97DD135 pKa = 3.74HH136 pKa = 7.1AGVLARR142 pKa = 11.84YY143 pKa = 7.77PKK145 pKa = 10.29FYY147 pKa = 9.18EE148 pKa = 4.14TLTLMARR155 pKa = 11.84PVRR158 pKa = 11.84TEE160 pKa = 3.56EE161 pKa = 4.0LQVILHH167 pKa = 6.82IGPTGLGKK175 pKa = 8.64TRR177 pKa = 11.84TVMDD181 pKa = 5.21LYY183 pKa = 11.24AADD186 pKa = 4.38PGLYY190 pKa = 8.3VTPLSNGTPWYY201 pKa = 10.04DD202 pKa = 3.56HH203 pKa = 7.01YY204 pKa = 11.43DD205 pKa = 3.3GHH207 pKa = 6.29HH208 pKa = 6.2TVLLDD213 pKa = 3.85DD214 pKa = 4.31FSGRR218 pKa = 11.84SSHH221 pKa = 6.05MSLVTLLRR229 pKa = 11.84LLDD232 pKa = 4.6RR233 pKa = 11.84YY234 pKa = 10.26PVMVPTKK241 pKa = 10.28GAHH244 pKa = 5.33TWWCPNTIYY253 pKa = 9.95VTTNILPSLWYY264 pKa = 8.8EE265 pKa = 3.44WGTRR269 pKa = 11.84GEE271 pKa = 4.37HH272 pKa = 5.23YY273 pKa = 9.9RR274 pKa = 11.84ALARR278 pKa = 11.84RR279 pKa = 11.84FTKK282 pKa = 10.43VVLFYY287 pKa = 11.05EE288 pKa = 4.76KK289 pKa = 10.71LHH291 pKa = 7.08ADD293 pKa = 3.58DD294 pKa = 5.5PGMVEE299 pKa = 5.86QDD301 pKa = 3.56ANWWKK306 pKa = 10.42EE307 pKa = 3.73NCPPEE312 pKa = 4.69AALLYY317 pKa = 10.38SQQ319 pKa = 5.02
MM1 pKa = 7.85AANQKK6 pKa = 9.52FKK8 pKa = 11.13NVCFTLNNPGEE19 pKa = 4.28DD20 pKa = 4.27PIPFDD25 pKa = 3.81VEE27 pKa = 3.76KK28 pKa = 10.0MHH30 pKa = 6.15YY31 pKa = 9.75LVYY34 pKa = 10.33QRR36 pKa = 11.84EE37 pKa = 4.2VGANGTEE44 pKa = 4.49HH45 pKa = 5.71YY46 pKa = 10.19QGYY49 pKa = 9.87CEE51 pKa = 5.9LINQTRR57 pKa = 11.84FNAVKK62 pKa = 10.14ALLGGGSVHH71 pKa = 6.44IEE73 pKa = 3.77RR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 10.69GSAQEE81 pKa = 3.41AAAYY85 pKa = 8.51CKK87 pKa = 10.34KK88 pKa = 10.66DD89 pKa = 3.45DD90 pKa = 3.77TRR92 pKa = 11.84APGASIQEE100 pKa = 4.29FGEE103 pKa = 4.09MHH105 pKa = 6.99ACTPGKK111 pKa = 10.64RR112 pKa = 11.84NDD114 pKa = 3.42LKK116 pKa = 11.47AFVDD120 pKa = 4.34DD121 pKa = 3.97VRR123 pKa = 11.84SGTKK127 pKa = 9.72RR128 pKa = 11.84KK129 pKa = 9.36RR130 pKa = 11.84DD131 pKa = 3.62LVEE134 pKa = 3.97DD135 pKa = 3.74HH136 pKa = 7.1AGVLARR142 pKa = 11.84YY143 pKa = 7.77PKK145 pKa = 10.29FYY147 pKa = 9.18EE148 pKa = 4.14TLTLMARR155 pKa = 11.84PVRR158 pKa = 11.84TEE160 pKa = 3.56EE161 pKa = 4.0LQVILHH167 pKa = 6.82IGPTGLGKK175 pKa = 8.64TRR177 pKa = 11.84TVMDD181 pKa = 5.21LYY183 pKa = 11.24AADD186 pKa = 4.38PGLYY190 pKa = 8.3VTPLSNGTPWYY201 pKa = 10.04DD202 pKa = 3.56HH203 pKa = 7.01YY204 pKa = 11.43DD205 pKa = 3.3GHH207 pKa = 6.29HH208 pKa = 6.2TVLLDD213 pKa = 3.85DD214 pKa = 4.31FSGRR218 pKa = 11.84SSHH221 pKa = 6.05MSLVTLLRR229 pKa = 11.84LLDD232 pKa = 4.6RR233 pKa = 11.84YY234 pKa = 10.26PVMVPTKK241 pKa = 10.28GAHH244 pKa = 5.33TWWCPNTIYY253 pKa = 9.95VTTNILPSLWYY264 pKa = 8.8EE265 pKa = 3.44WGTRR269 pKa = 11.84GEE271 pKa = 4.37HH272 pKa = 5.23YY273 pKa = 9.9RR274 pKa = 11.84ALARR278 pKa = 11.84RR279 pKa = 11.84FTKK282 pKa = 10.43VVLFYY287 pKa = 11.05EE288 pKa = 4.76KK289 pKa = 10.71LHH291 pKa = 7.08ADD293 pKa = 3.58DD294 pKa = 5.5PGMVEE299 pKa = 5.86QDD301 pKa = 3.56ANWWKK306 pKa = 10.42EE307 pKa = 3.73NCPPEE312 pKa = 4.69AALLYY317 pKa = 10.38SQQ319 pKa = 5.02
Molecular weight: 36.26 kDa
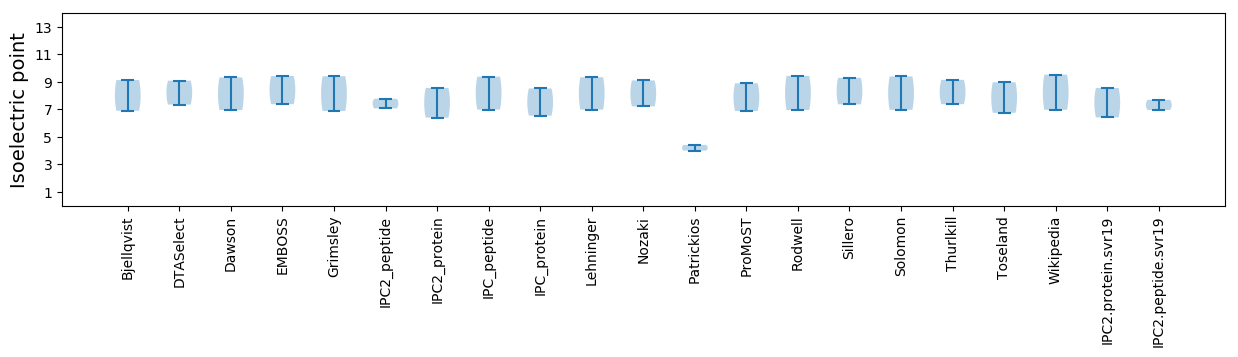
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IM93|A0A0C5IM93_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 5 OX=1618256 PE=3 SV=1
MM1 pKa = 7.36SASGYY6 pKa = 10.15YY7 pKa = 10.04SGNKK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 10.12RR14 pKa = 11.84SDD16 pKa = 3.59FSADD20 pKa = 3.18AQALAVARR28 pKa = 11.84PSFKK32 pKa = 10.16GYY34 pKa = 9.64KK35 pKa = 9.14RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.96EE40 pKa = 3.63FVPGQDD46 pKa = 2.63RR47 pKa = 11.84TGGYY51 pKa = 10.16YY52 pKa = 10.22RR53 pKa = 11.84FGKK56 pKa = 7.96TAMGPGMGEE65 pKa = 3.8MKK67 pKa = 10.66FFDD70 pKa = 3.74TDD72 pKa = 3.17ISEE75 pKa = 4.2LTILDD80 pKa = 3.57GGIVKK85 pKa = 10.66ASMNNVPQGTGEE97 pKa = 3.98SQRR100 pKa = 11.84IGRR103 pKa = 11.84KK104 pKa = 9.13CVVKK108 pKa = 10.49SIYY111 pKa = 10.74LKK113 pKa = 10.79FSVNLPQLEE122 pKa = 4.41NEE124 pKa = 4.52VNMKK128 pKa = 10.56NPDD131 pKa = 3.3QVRR134 pKa = 11.84VILYY138 pKa = 10.1LDD140 pKa = 3.47KK141 pKa = 10.89QANGEE146 pKa = 4.32TTTLGSILEE155 pKa = 4.36GDD157 pKa = 3.88PKK159 pKa = 10.6ISSFRR164 pKa = 11.84NLSNMNRR171 pKa = 11.84YY172 pKa = 7.5TILMDD177 pKa = 3.55KK178 pKa = 10.4VYY180 pKa = 11.1DD181 pKa = 3.65MNYY184 pKa = 9.7EE185 pKa = 4.15SVAAEE190 pKa = 3.92TDD192 pKa = 3.36QGDD195 pKa = 3.33VRR197 pKa = 11.84YY198 pKa = 11.0SMVGKK203 pKa = 7.7TCYY206 pKa = 9.41VSEE209 pKa = 4.01YY210 pKa = 9.98RR211 pKa = 11.84KK212 pKa = 10.53LNIPIEE218 pKa = 4.39FSGTMGAIDD227 pKa = 4.73EE228 pKa = 4.77IGSNNIGLGYY238 pKa = 10.16ISANNVAQIVGQTRR252 pKa = 11.84IRR254 pKa = 11.84YY255 pKa = 9.39ADD257 pKa = 3.13AA258 pKa = 5.34
MM1 pKa = 7.36SASGYY6 pKa = 10.15YY7 pKa = 10.04SGNKK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 10.12RR14 pKa = 11.84SDD16 pKa = 3.59FSADD20 pKa = 3.18AQALAVARR28 pKa = 11.84PSFKK32 pKa = 10.16GYY34 pKa = 9.64KK35 pKa = 9.14RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.96EE40 pKa = 3.63FVPGQDD46 pKa = 2.63RR47 pKa = 11.84TGGYY51 pKa = 10.16YY52 pKa = 10.22RR53 pKa = 11.84FGKK56 pKa = 7.96TAMGPGMGEE65 pKa = 3.8MKK67 pKa = 10.66FFDD70 pKa = 3.74TDD72 pKa = 3.17ISEE75 pKa = 4.2LTILDD80 pKa = 3.57GGIVKK85 pKa = 10.66ASMNNVPQGTGEE97 pKa = 3.98SQRR100 pKa = 11.84IGRR103 pKa = 11.84KK104 pKa = 9.13CVVKK108 pKa = 10.49SIYY111 pKa = 10.74LKK113 pKa = 10.79FSVNLPQLEE122 pKa = 4.41NEE124 pKa = 4.52VNMKK128 pKa = 10.56NPDD131 pKa = 3.3QVRR134 pKa = 11.84VILYY138 pKa = 10.1LDD140 pKa = 3.47KK141 pKa = 10.89QANGEE146 pKa = 4.32TTTLGSILEE155 pKa = 4.36GDD157 pKa = 3.88PKK159 pKa = 10.6ISSFRR164 pKa = 11.84NLSNMNRR171 pKa = 11.84YY172 pKa = 7.5TILMDD177 pKa = 3.55KK178 pKa = 10.4VYY180 pKa = 11.1DD181 pKa = 3.65MNYY184 pKa = 9.7EE185 pKa = 4.15SVAAEE190 pKa = 3.92TDD192 pKa = 3.36QGDD195 pKa = 3.33VRR197 pKa = 11.84YY198 pKa = 11.0SMVGKK203 pKa = 7.7TCYY206 pKa = 9.41VSEE209 pKa = 4.01YY210 pKa = 9.98RR211 pKa = 11.84KK212 pKa = 10.53LNIPIEE218 pKa = 4.39FSGTMGAIDD227 pKa = 4.73EE228 pKa = 4.77IGSNNIGLGYY238 pKa = 10.16ISANNVAQIVGQTRR252 pKa = 11.84IRR254 pKa = 11.84YY255 pKa = 9.39ADD257 pKa = 3.13AA258 pKa = 5.34
Molecular weight: 28.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
577 |
258 |
319 |
288.5 |
32.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.106 ± 0.546 | 1.386 ± 0.369 |
5.893 ± 0.047 | 5.546 ± 0.306 |
3.293 ± 0.118 | 8.666 ± 0.853 |
2.253 ± 1.361 | 4.333 ± 1.363 |
5.893 ± 0.421 | 7.626 ± 1.328 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.293 ± 0.586 | 5.199 ± 0.839 |
4.506 ± 0.849 | 3.293 ± 0.352 |
6.412 ± 0.107 | 5.546 ± 1.566 |
6.239 ± 0.725 | 6.759 ± 0.103 |
1.213 ± 0.733 | 5.546 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
