
Tioman virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Pararubulavirus; Tioman pararubulavirus
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
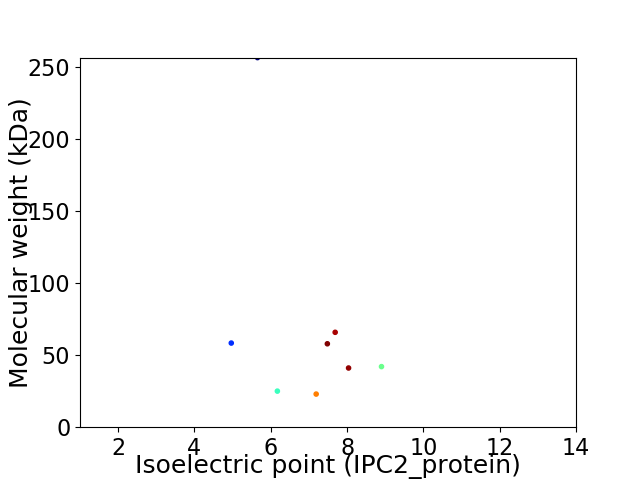
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8JVB0|Q8JVB0_9MONO Isoform of Q8JVB1 W protein OS=Tioman virus OX=162013 GN=P PE=4 SV=1
MM1 pKa = 7.4SSVFRR6 pKa = 11.84AFEE9 pKa = 4.3LFTLEE14 pKa = 5.1QEE16 pKa = 4.36QQEE19 pKa = 4.48LGNDD23 pKa = 3.2IEE25 pKa = 4.96IPPEE29 pKa = 3.91TLRR32 pKa = 11.84SNIKK36 pKa = 9.81VCILNSQDD44 pKa = 3.01PQTRR48 pKa = 11.84HH49 pKa = 7.0DD50 pKa = 4.01MMCFCLRR57 pKa = 11.84LIASNSARR65 pKa = 11.84AAHH68 pKa = 5.96KK69 pKa = 9.34TGAILTLLNLPTAMMQNHH87 pKa = 5.88IRR89 pKa = 11.84IADD92 pKa = 4.24RR93 pKa = 11.84SPDD96 pKa = 3.38ADD98 pKa = 3.22IEE100 pKa = 4.36RR101 pKa = 11.84IEE103 pKa = 4.01VDD105 pKa = 3.08GFEE108 pKa = 4.55PGTYY112 pKa = 9.65RR113 pKa = 11.84LRR115 pKa = 11.84PNARR119 pKa = 11.84TPLTNGEE126 pKa = 3.93ITALSLMANDD136 pKa = 5.85LPDD139 pKa = 3.74TYY141 pKa = 11.46TNDD144 pKa = 3.58TPFTNHH150 pKa = 6.12RR151 pKa = 11.84AEE153 pKa = 5.09GEE155 pKa = 4.19NCDD158 pKa = 3.69EE159 pKa = 4.45TEE161 pKa = 3.87QFLNAIYY168 pKa = 10.52SVLVQLWVTVCKK180 pKa = 10.64CMTAHH185 pKa = 6.77DD186 pKa = 4.33QPTGSDD192 pKa = 3.06EE193 pKa = 4.13RR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.17YY199 pKa = 8.02QQQGRR204 pKa = 11.84LDD206 pKa = 3.35QKK208 pKa = 11.1YY209 pKa = 10.28ALQPEE214 pKa = 4.95LRR216 pKa = 11.84RR217 pKa = 11.84QIQACIRR224 pKa = 11.84GSLTIRR230 pKa = 11.84QFLTYY235 pKa = 10.1EE236 pKa = 3.87LQTARR241 pKa = 11.84KK242 pKa = 8.48QGAITGRR249 pKa = 11.84YY250 pKa = 7.34YY251 pKa = 11.77AMVGDD256 pKa = 3.52IGKK259 pKa = 10.43YY260 pKa = 9.05IDD262 pKa = 3.75NAGMSAFFMTMRR274 pKa = 11.84FALGTRR280 pKa = 11.84WPPLALSAFSGEE292 pKa = 4.03LLKK295 pKa = 11.09LKK297 pKa = 10.68SLMQLYY303 pKa = 10.58RR304 pKa = 11.84NLGEE308 pKa = 4.16KK309 pKa = 9.58ARR311 pKa = 11.84YY312 pKa = 7.03MALLEE317 pKa = 4.25MPEE320 pKa = 4.02MMEE323 pKa = 4.34FAPANYY329 pKa = 8.35PLCYY333 pKa = 9.75SYY335 pKa = 11.89AMGIGSVQDD344 pKa = 3.3PMMRR348 pKa = 11.84NYY350 pKa = 9.81TFARR354 pKa = 11.84PFLNPAYY361 pKa = 9.55FQLGVEE367 pKa = 4.29TANRR371 pKa = 11.84QQGSVDD377 pKa = 2.85KK378 pKa = 11.56SMAEE382 pKa = 3.8EE383 pKa = 4.3LGLTEE388 pKa = 4.13EE389 pKa = 4.56EE390 pKa = 4.2RR391 pKa = 11.84RR392 pKa = 11.84DD393 pKa = 3.35MSATVTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GAGQAQDD412 pKa = 4.28MINIMGARR420 pKa = 11.84QAAGGRR426 pKa = 11.84GAQGRR431 pKa = 11.84ALRR434 pKa = 11.84IVEE437 pKa = 4.19EE438 pKa = 5.16DD439 pKa = 3.21EE440 pKa = 4.36TTDD443 pKa = 4.67DD444 pKa = 5.07EE445 pKa = 5.44SVDD448 pKa = 3.87DD449 pKa = 4.64QADD452 pKa = 4.39DD453 pKa = 3.44IQGRR457 pKa = 11.84PLPPVPAPIRR467 pKa = 11.84EE468 pKa = 4.21IDD470 pKa = 3.14WEE472 pKa = 3.98ARR474 pKa = 11.84IAEE477 pKa = 4.32IEE479 pKa = 4.17EE480 pKa = 3.71QGQRR484 pKa = 11.84MRR486 pKa = 11.84DD487 pKa = 3.33QGVGRR492 pKa = 11.84LVDD495 pKa = 3.54NQAPNVQNTRR505 pKa = 11.84QNTQDD510 pKa = 3.33NQALLDD516 pKa = 4.0LDD518 pKa = 3.67II519 pKa = 6.3
MM1 pKa = 7.4SSVFRR6 pKa = 11.84AFEE9 pKa = 4.3LFTLEE14 pKa = 5.1QEE16 pKa = 4.36QQEE19 pKa = 4.48LGNDD23 pKa = 3.2IEE25 pKa = 4.96IPPEE29 pKa = 3.91TLRR32 pKa = 11.84SNIKK36 pKa = 9.81VCILNSQDD44 pKa = 3.01PQTRR48 pKa = 11.84HH49 pKa = 7.0DD50 pKa = 4.01MMCFCLRR57 pKa = 11.84LIASNSARR65 pKa = 11.84AAHH68 pKa = 5.96KK69 pKa = 9.34TGAILTLLNLPTAMMQNHH87 pKa = 5.88IRR89 pKa = 11.84IADD92 pKa = 4.24RR93 pKa = 11.84SPDD96 pKa = 3.38ADD98 pKa = 3.22IEE100 pKa = 4.36RR101 pKa = 11.84IEE103 pKa = 4.01VDD105 pKa = 3.08GFEE108 pKa = 4.55PGTYY112 pKa = 9.65RR113 pKa = 11.84LRR115 pKa = 11.84PNARR119 pKa = 11.84TPLTNGEE126 pKa = 3.93ITALSLMANDD136 pKa = 5.85LPDD139 pKa = 3.74TYY141 pKa = 11.46TNDD144 pKa = 3.58TPFTNHH150 pKa = 6.12RR151 pKa = 11.84AEE153 pKa = 5.09GEE155 pKa = 4.19NCDD158 pKa = 3.69EE159 pKa = 4.45TEE161 pKa = 3.87QFLNAIYY168 pKa = 10.52SVLVQLWVTVCKK180 pKa = 10.64CMTAHH185 pKa = 6.77DD186 pKa = 4.33QPTGSDD192 pKa = 3.06EE193 pKa = 4.13RR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.17YY199 pKa = 8.02QQQGRR204 pKa = 11.84LDD206 pKa = 3.35QKK208 pKa = 11.1YY209 pKa = 10.28ALQPEE214 pKa = 4.95LRR216 pKa = 11.84RR217 pKa = 11.84QIQACIRR224 pKa = 11.84GSLTIRR230 pKa = 11.84QFLTYY235 pKa = 10.1EE236 pKa = 3.87LQTARR241 pKa = 11.84KK242 pKa = 8.48QGAITGRR249 pKa = 11.84YY250 pKa = 7.34YY251 pKa = 11.77AMVGDD256 pKa = 3.52IGKK259 pKa = 10.43YY260 pKa = 9.05IDD262 pKa = 3.75NAGMSAFFMTMRR274 pKa = 11.84FALGTRR280 pKa = 11.84WPPLALSAFSGEE292 pKa = 4.03LLKK295 pKa = 11.09LKK297 pKa = 10.68SLMQLYY303 pKa = 10.58RR304 pKa = 11.84NLGEE308 pKa = 4.16KK309 pKa = 9.58ARR311 pKa = 11.84YY312 pKa = 7.03MALLEE317 pKa = 4.25MPEE320 pKa = 4.02MMEE323 pKa = 4.34FAPANYY329 pKa = 8.35PLCYY333 pKa = 9.75SYY335 pKa = 11.89AMGIGSVQDD344 pKa = 3.3PMMRR348 pKa = 11.84NYY350 pKa = 9.81TFARR354 pKa = 11.84PFLNPAYY361 pKa = 9.55FQLGVEE367 pKa = 4.29TANRR371 pKa = 11.84QQGSVDD377 pKa = 2.85KK378 pKa = 11.56SMAEE382 pKa = 3.8EE383 pKa = 4.3LGLTEE388 pKa = 4.13EE389 pKa = 4.56EE390 pKa = 4.2RR391 pKa = 11.84RR392 pKa = 11.84DD393 pKa = 3.35MSATVTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GAGQAQDD412 pKa = 4.28MINIMGARR420 pKa = 11.84QAAGGRR426 pKa = 11.84GAQGRR431 pKa = 11.84ALRR434 pKa = 11.84IVEE437 pKa = 4.19EE438 pKa = 5.16DD439 pKa = 3.21EE440 pKa = 4.36TTDD443 pKa = 4.67DD444 pKa = 5.07EE445 pKa = 5.44SVDD448 pKa = 3.87DD449 pKa = 4.64QADD452 pKa = 4.39DD453 pKa = 3.44IQGRR457 pKa = 11.84PLPPVPAPIRR467 pKa = 11.84EE468 pKa = 4.21IDD470 pKa = 3.14WEE472 pKa = 3.98ARR474 pKa = 11.84IAEE477 pKa = 4.32IEE479 pKa = 4.17EE480 pKa = 3.71QGQRR484 pKa = 11.84MRR486 pKa = 11.84DD487 pKa = 3.33QGVGRR492 pKa = 11.84LVDD495 pKa = 3.54NQAPNVQNTRR505 pKa = 11.84QNTQDD510 pKa = 3.33NQALLDD516 pKa = 4.0LDD518 pKa = 3.67II519 pKa = 6.3
Molecular weight: 58.44 kDa
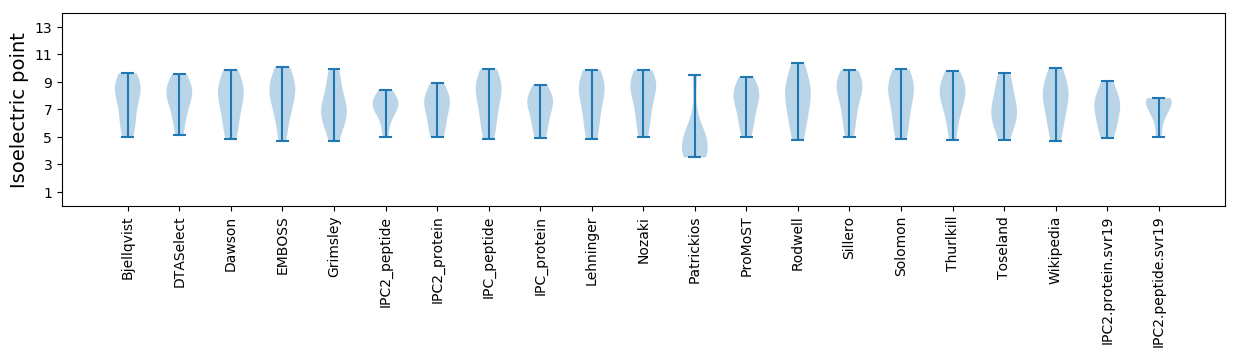
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8JVB1|Q8JVB1_9MONO Phosphoprotein OS=Tioman virus OX=162013 GN=P PE=3 SV=1
MM1 pKa = 8.01ALRR4 pKa = 11.84QATIPINVDD13 pKa = 3.2TEE15 pKa = 4.48SEE17 pKa = 4.29KK18 pKa = 11.46NNLNPFPIVPITKK31 pKa = 9.82EE32 pKa = 4.01DD33 pKa = 3.52GSPTGRR39 pKa = 11.84LVKK42 pKa = 9.79QLRR45 pKa = 11.84IKK47 pKa = 10.84NLTPRR52 pKa = 11.84GSTEE56 pKa = 4.25LPLTFINTYY65 pKa = 10.73GFIKK69 pKa = 10.48PLMTYY74 pKa = 8.72TEE76 pKa = 4.46FFSTHH81 pKa = 6.0LRR83 pKa = 11.84TPSCLTACMIPFGAGPFIEE102 pKa = 4.56NPHH105 pKa = 6.88RR106 pKa = 11.84ILDD109 pKa = 3.54KK110 pKa = 10.56CDD112 pKa = 3.0RR113 pKa = 11.84VNIVVRR119 pKa = 11.84KK120 pKa = 7.83SASVKK125 pKa = 10.25EE126 pKa = 4.11EE127 pKa = 3.75IIFDD131 pKa = 3.88VRR133 pKa = 11.84RR134 pKa = 11.84LPPLFNRR141 pKa = 11.84HH142 pKa = 5.38QISGNRR148 pKa = 11.84LICVPSEE155 pKa = 4.41KK156 pKa = 10.2YY157 pKa = 10.28VKK159 pKa = 10.73SPGKK163 pKa = 9.74MIAGTDD169 pKa = 3.53YY170 pKa = 10.53AYY172 pKa = 10.85QIAFVSLTFCPEE184 pKa = 3.79SQKK187 pKa = 10.37FRR189 pKa = 11.84VARR192 pKa = 11.84PLQTIRR198 pKa = 11.84SPIMRR203 pKa = 11.84SVQLEE208 pKa = 4.42VILKK212 pKa = 9.46IDD214 pKa = 3.71CAANSPLKK222 pKa = 10.32RR223 pKa = 11.84FLIISPNSKK232 pKa = 10.66DD233 pKa = 3.23CFASVWFHH241 pKa = 6.1ICNLYY246 pKa = 10.45RR247 pKa = 11.84GNKK250 pKa = 8.09PFKK253 pKa = 10.56AYY255 pKa = 10.46DD256 pKa = 3.37DD257 pKa = 4.34TYY259 pKa = 11.28FSQKK263 pKa = 8.99CRR265 pKa = 11.84AMQLEE270 pKa = 4.42CGIVDD275 pKa = 3.09MWGPTLVVKK284 pKa = 10.71AHH286 pKa = 5.64GKK288 pKa = 7.29IPKK291 pKa = 8.05MARR294 pKa = 11.84PFFSSKK300 pKa = 9.41GWSCHH305 pKa = 5.21AFADD309 pKa = 4.83AAPTLAKK316 pKa = 10.59ALWSVGAQITQVNAILQPSDD336 pKa = 3.11LHH338 pKa = 7.41QLVQISDD345 pKa = 4.21VIWPKK350 pKa = 10.84VKK352 pKa = 10.27LDD354 pKa = 3.94EE355 pKa = 5.62KK356 pKa = 9.24IQSYY360 pKa = 10.08AAAKK364 pKa = 8.17WNPFKK369 pKa = 11.08KK370 pKa = 9.33STNN373 pKa = 3.45
MM1 pKa = 8.01ALRR4 pKa = 11.84QATIPINVDD13 pKa = 3.2TEE15 pKa = 4.48SEE17 pKa = 4.29KK18 pKa = 11.46NNLNPFPIVPITKK31 pKa = 9.82EE32 pKa = 4.01DD33 pKa = 3.52GSPTGRR39 pKa = 11.84LVKK42 pKa = 9.79QLRR45 pKa = 11.84IKK47 pKa = 10.84NLTPRR52 pKa = 11.84GSTEE56 pKa = 4.25LPLTFINTYY65 pKa = 10.73GFIKK69 pKa = 10.48PLMTYY74 pKa = 8.72TEE76 pKa = 4.46FFSTHH81 pKa = 6.0LRR83 pKa = 11.84TPSCLTACMIPFGAGPFIEE102 pKa = 4.56NPHH105 pKa = 6.88RR106 pKa = 11.84ILDD109 pKa = 3.54KK110 pKa = 10.56CDD112 pKa = 3.0RR113 pKa = 11.84VNIVVRR119 pKa = 11.84KK120 pKa = 7.83SASVKK125 pKa = 10.25EE126 pKa = 4.11EE127 pKa = 3.75IIFDD131 pKa = 3.88VRR133 pKa = 11.84RR134 pKa = 11.84LPPLFNRR141 pKa = 11.84HH142 pKa = 5.38QISGNRR148 pKa = 11.84LICVPSEE155 pKa = 4.41KK156 pKa = 10.2YY157 pKa = 10.28VKK159 pKa = 10.73SPGKK163 pKa = 9.74MIAGTDD169 pKa = 3.53YY170 pKa = 10.53AYY172 pKa = 10.85QIAFVSLTFCPEE184 pKa = 3.79SQKK187 pKa = 10.37FRR189 pKa = 11.84VARR192 pKa = 11.84PLQTIRR198 pKa = 11.84SPIMRR203 pKa = 11.84SVQLEE208 pKa = 4.42VILKK212 pKa = 9.46IDD214 pKa = 3.71CAANSPLKK222 pKa = 10.32RR223 pKa = 11.84FLIISPNSKK232 pKa = 10.66DD233 pKa = 3.23CFASVWFHH241 pKa = 6.1ICNLYY246 pKa = 10.45RR247 pKa = 11.84GNKK250 pKa = 8.09PFKK253 pKa = 10.56AYY255 pKa = 10.46DD256 pKa = 3.37DD257 pKa = 4.34TYY259 pKa = 11.28FSQKK263 pKa = 8.99CRR265 pKa = 11.84AMQLEE270 pKa = 4.42CGIVDD275 pKa = 3.09MWGPTLVVKK284 pKa = 10.71AHH286 pKa = 5.64GKK288 pKa = 7.29IPKK291 pKa = 8.05MARR294 pKa = 11.84PFFSSKK300 pKa = 9.41GWSCHH305 pKa = 5.21AFADD309 pKa = 4.83AAPTLAKK316 pKa = 10.59ALWSVGAQITQVNAILQPSDD336 pKa = 3.11LHH338 pKa = 7.41QLVQISDD345 pKa = 4.21VIWPKK350 pKa = 10.84VKK352 pKa = 10.27LDD354 pKa = 3.94EE355 pKa = 5.62KK356 pKa = 9.24IQSYY360 pKa = 10.08AAAKK364 pKa = 8.17WNPFKK369 pKa = 11.08KK370 pKa = 9.33STNN373 pKa = 3.45
Molecular weight: 42.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5118 |
211 |
2271 |
639.8 |
71.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.585 ± 0.829 | 2.188 ± 0.252 |
5.1 ± 0.589 | 4.982 ± 0.584 |
3.243 ± 0.389 | 5.275 ± 0.566 |
1.837 ± 0.221 | 7.64 ± 0.554 |
4.787 ± 0.565 | 10.746 ± 1.234 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.349 | 5.178 ± 0.443 |
5.569 ± 0.753 | 4.572 ± 0.522 |
5.178 ± 0.509 | 8.304 ± 0.618 |
6.643 ± 0.241 | 5.686 ± 0.484 |
1.055 ± 0.152 | 3.029 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
