
Erysiphe necator mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I5ARE7|A0A2I5ARE7_9VIRU RNA-dependent RNA polymerase OS=Erysiphe necator mitovirus 1 OX=2052561 PE=4 SV=1
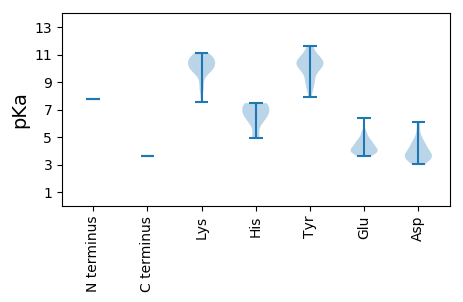
MM1 pKa = 7.74AGEE4 pKa = 4.71SCCSVQKK11 pKa = 11.02DD12 pKa = 3.41HH13 pKa = 7.36VLYY16 pKa = 10.18LWGWIEE22 pKa = 3.93TSLKK26 pKa = 10.71HH27 pKa = 6.38KK28 pKa = 10.32GLSHH32 pKa = 7.34TILSLKK38 pKa = 8.14TMRR41 pKa = 11.84NCVTKK46 pKa = 10.27YY47 pKa = 10.13IAGEE51 pKa = 3.79PVKK54 pKa = 10.73VVEE57 pKa = 4.86IRR59 pKa = 11.84LSLTKK64 pKa = 10.61DD65 pKa = 4.32GIPKK69 pKa = 10.42ALGPLVPIIRR79 pKa = 11.84KK80 pKa = 9.46LEE82 pKa = 4.2PQDD85 pKa = 3.54LRR87 pKa = 11.84IVLTLLSVGKK97 pKa = 9.96LYY99 pKa = 10.51PGDD102 pKa = 3.45GTVNTKK108 pKa = 10.66SITDD112 pKa = 3.8PFSGSMSKK120 pKa = 10.04LHH122 pKa = 6.76EE123 pKa = 4.09YY124 pKa = 10.66ALRR127 pKa = 11.84GSFILEE133 pKa = 3.69NLFKK137 pKa = 11.12NQLTQLDD144 pKa = 4.41IEE146 pKa = 4.9QSSNWNAPHH155 pKa = 7.46LSTKK159 pKa = 10.06KK160 pKa = 10.8GPIGHH165 pKa = 7.4ALGTSMIEE173 pKa = 4.6LGHH176 pKa = 7.45LPDD179 pKa = 4.63KK180 pKa = 11.1LLVQIIKK187 pKa = 10.75LGGLKK192 pKa = 10.06LRR194 pKa = 11.84NYY196 pKa = 9.08ILSIIKK202 pKa = 8.66MARR205 pKa = 11.84TISKK209 pKa = 10.28FEE211 pKa = 4.06NSKK214 pKa = 10.88KK215 pKa = 10.18PIPKK219 pKa = 9.64VYY221 pKa = 10.19RR222 pKa = 11.84KK223 pKa = 10.24LSIVKK228 pKa = 10.33DD229 pKa = 3.63KK230 pKa = 10.65EE231 pKa = 4.26LKK233 pKa = 9.98NRR235 pKa = 11.84PIAIFDD241 pKa = 3.62YY242 pKa = 10.11WSQSALKK249 pKa = 8.77PLHH252 pKa = 6.78DD253 pKa = 3.91SVMNLLRR260 pKa = 11.84TFKK263 pKa = 10.62TDD265 pKa = 3.08LTFNQEE271 pKa = 3.73GCEE274 pKa = 4.22EE275 pKa = 4.24IPSLIGPQAKK285 pKa = 9.98LFSLDD290 pKa = 3.65LSSATDD296 pKa = 3.69RR297 pKa = 11.84FPVEE301 pKa = 3.75FQEE304 pKa = 4.79IIVSRR309 pKa = 11.84LIGQEE314 pKa = 3.62RR315 pKa = 11.84AEE317 pKa = 3.84AWRR320 pKa = 11.84YY321 pKa = 10.33IMTEE325 pKa = 4.06FPFNTPEE332 pKa = 4.49GDD334 pKa = 3.61SVKK337 pKa = 10.75YY338 pKa = 8.79NTGQPMGGYY347 pKa = 9.81SSWCIFTLCHH357 pKa = 6.18HH358 pKa = 6.86LVVQIAYY365 pKa = 10.26NEE367 pKa = 4.15AYY369 pKa = 9.23NRR371 pKa = 11.84NWSRR375 pKa = 11.84GARR378 pKa = 11.84SGLPITPKK386 pKa = 10.42LFEE389 pKa = 5.29DD390 pKa = 3.97YY391 pKa = 10.53RR392 pKa = 11.84ILGDD396 pKa = 6.0DD397 pKa = 3.21IVIWDD402 pKa = 3.62EE403 pKa = 3.96MVARR407 pKa = 11.84SYY409 pKa = 11.31KK410 pKa = 10.7SIMADD415 pKa = 3.15LGVEE419 pKa = 4.0ISPFKK424 pKa = 10.31SHH426 pKa = 6.21EE427 pKa = 4.36SYY429 pKa = 8.76HH430 pKa = 4.93TFEE433 pKa = 3.81MAKK436 pKa = 9.65RR437 pKa = 11.84WFHH440 pKa = 6.73KK441 pKa = 9.62GTEE444 pKa = 4.02ITGFPLNNLLEE455 pKa = 4.68AKK457 pKa = 10.08SNPWGIGKK465 pKa = 9.7ALSDD469 pKa = 4.57GIWRR473 pKa = 11.84GWITKK478 pKa = 9.91SNQGVPGNILIRR490 pKa = 11.84RR491 pKa = 11.84LLYY494 pKa = 10.58SLGWSKK500 pKa = 11.0RR501 pKa = 11.84SINNRR506 pKa = 11.84EE507 pKa = 3.62EE508 pKa = 4.1SVRR511 pKa = 11.84DD512 pKa = 3.29SFLFFNFSVTRR523 pKa = 11.84KK524 pKa = 7.52TDD526 pKa = 3.03WVGEE530 pKa = 3.92RR531 pKa = 11.84SVRR534 pKa = 11.84EE535 pKa = 3.82FVSRR539 pKa = 11.84HH540 pKa = 4.99GPGQTMYY547 pKa = 7.93TTQSARR553 pKa = 11.84QEE555 pKa = 3.85LAEE558 pKa = 4.07VVRR561 pKa = 11.84RR562 pKa = 11.84IRR564 pKa = 11.84VKK566 pKa = 11.01GLLSLLNKK574 pKa = 9.72GQEE577 pKa = 4.37LIEE580 pKa = 4.2KK581 pKa = 9.97RR582 pKa = 11.84NTAFGDD588 pKa = 3.62YY589 pKa = 10.39LRR591 pKa = 11.84SLKK594 pKa = 10.82DD595 pKa = 3.32CGLPPTAFSLIEE607 pKa = 4.54SIPVFQVLDD616 pKa = 3.98RR617 pKa = 11.84NQDD620 pKa = 3.91LIKK623 pKa = 10.6QAVIDD628 pKa = 3.97LEE630 pKa = 4.54GTNEE634 pKa = 3.91EE635 pKa = 4.49YY636 pKa = 10.96ALSLDD641 pKa = 3.6EE642 pKa = 4.68SLSIGADD649 pKa = 3.2LVVPDD654 pKa = 5.03PDD656 pKa = 4.15YY657 pKa = 11.65IMSEE661 pKa = 3.92RR662 pKa = 11.84KK663 pKa = 9.1SRR665 pKa = 11.84LIIKK669 pKa = 10.05QNAKK673 pKa = 8.54TLATLLCAFEE683 pKa = 5.35DD684 pKa = 3.98PLIIIDD690 pKa = 6.08DD691 pKa = 4.7DD692 pKa = 4.46DD693 pKa = 4.62QDD695 pKa = 3.81TCIEE699 pKa = 4.48DD700 pKa = 5.15DD701 pKa = 4.28EE702 pKa = 6.42DD703 pKa = 4.56SEE705 pKa = 4.93PDD707 pKa = 3.44SLFQGLYY714 pKa = 10.49DD715 pKa = 5.45EE716 pKa = 5.11EE717 pKa = 5.47PDD719 pKa = 3.29WSAMM723 pKa = 3.64
MM1 pKa = 7.74AGEE4 pKa = 4.71SCCSVQKK11 pKa = 11.02DD12 pKa = 3.41HH13 pKa = 7.36VLYY16 pKa = 10.18LWGWIEE22 pKa = 3.93TSLKK26 pKa = 10.71HH27 pKa = 6.38KK28 pKa = 10.32GLSHH32 pKa = 7.34TILSLKK38 pKa = 8.14TMRR41 pKa = 11.84NCVTKK46 pKa = 10.27YY47 pKa = 10.13IAGEE51 pKa = 3.79PVKK54 pKa = 10.73VVEE57 pKa = 4.86IRR59 pKa = 11.84LSLTKK64 pKa = 10.61DD65 pKa = 4.32GIPKK69 pKa = 10.42ALGPLVPIIRR79 pKa = 11.84KK80 pKa = 9.46LEE82 pKa = 4.2PQDD85 pKa = 3.54LRR87 pKa = 11.84IVLTLLSVGKK97 pKa = 9.96LYY99 pKa = 10.51PGDD102 pKa = 3.45GTVNTKK108 pKa = 10.66SITDD112 pKa = 3.8PFSGSMSKK120 pKa = 10.04LHH122 pKa = 6.76EE123 pKa = 4.09YY124 pKa = 10.66ALRR127 pKa = 11.84GSFILEE133 pKa = 3.69NLFKK137 pKa = 11.12NQLTQLDD144 pKa = 4.41IEE146 pKa = 4.9QSSNWNAPHH155 pKa = 7.46LSTKK159 pKa = 10.06KK160 pKa = 10.8GPIGHH165 pKa = 7.4ALGTSMIEE173 pKa = 4.6LGHH176 pKa = 7.45LPDD179 pKa = 4.63KK180 pKa = 11.1LLVQIIKK187 pKa = 10.75LGGLKK192 pKa = 10.06LRR194 pKa = 11.84NYY196 pKa = 9.08ILSIIKK202 pKa = 8.66MARR205 pKa = 11.84TISKK209 pKa = 10.28FEE211 pKa = 4.06NSKK214 pKa = 10.88KK215 pKa = 10.18PIPKK219 pKa = 9.64VYY221 pKa = 10.19RR222 pKa = 11.84KK223 pKa = 10.24LSIVKK228 pKa = 10.33DD229 pKa = 3.63KK230 pKa = 10.65EE231 pKa = 4.26LKK233 pKa = 9.98NRR235 pKa = 11.84PIAIFDD241 pKa = 3.62YY242 pKa = 10.11WSQSALKK249 pKa = 8.77PLHH252 pKa = 6.78DD253 pKa = 3.91SVMNLLRR260 pKa = 11.84TFKK263 pKa = 10.62TDD265 pKa = 3.08LTFNQEE271 pKa = 3.73GCEE274 pKa = 4.22EE275 pKa = 4.24IPSLIGPQAKK285 pKa = 9.98LFSLDD290 pKa = 3.65LSSATDD296 pKa = 3.69RR297 pKa = 11.84FPVEE301 pKa = 3.75FQEE304 pKa = 4.79IIVSRR309 pKa = 11.84LIGQEE314 pKa = 3.62RR315 pKa = 11.84AEE317 pKa = 3.84AWRR320 pKa = 11.84YY321 pKa = 10.33IMTEE325 pKa = 4.06FPFNTPEE332 pKa = 4.49GDD334 pKa = 3.61SVKK337 pKa = 10.75YY338 pKa = 8.79NTGQPMGGYY347 pKa = 9.81SSWCIFTLCHH357 pKa = 6.18HH358 pKa = 6.86LVVQIAYY365 pKa = 10.26NEE367 pKa = 4.15AYY369 pKa = 9.23NRR371 pKa = 11.84NWSRR375 pKa = 11.84GARR378 pKa = 11.84SGLPITPKK386 pKa = 10.42LFEE389 pKa = 5.29DD390 pKa = 3.97YY391 pKa = 10.53RR392 pKa = 11.84ILGDD396 pKa = 6.0DD397 pKa = 3.21IVIWDD402 pKa = 3.62EE403 pKa = 3.96MVARR407 pKa = 11.84SYY409 pKa = 11.31KK410 pKa = 10.7SIMADD415 pKa = 3.15LGVEE419 pKa = 4.0ISPFKK424 pKa = 10.31SHH426 pKa = 6.21EE427 pKa = 4.36SYY429 pKa = 8.76HH430 pKa = 4.93TFEE433 pKa = 3.81MAKK436 pKa = 9.65RR437 pKa = 11.84WFHH440 pKa = 6.73KK441 pKa = 9.62GTEE444 pKa = 4.02ITGFPLNNLLEE455 pKa = 4.68AKK457 pKa = 10.08SNPWGIGKK465 pKa = 9.7ALSDD469 pKa = 4.57GIWRR473 pKa = 11.84GWITKK478 pKa = 9.91SNQGVPGNILIRR490 pKa = 11.84RR491 pKa = 11.84LLYY494 pKa = 10.58SLGWSKK500 pKa = 11.0RR501 pKa = 11.84SINNRR506 pKa = 11.84EE507 pKa = 3.62EE508 pKa = 4.1SVRR511 pKa = 11.84DD512 pKa = 3.29SFLFFNFSVTRR523 pKa = 11.84KK524 pKa = 7.52TDD526 pKa = 3.03WVGEE530 pKa = 3.92RR531 pKa = 11.84SVRR534 pKa = 11.84EE535 pKa = 3.82FVSRR539 pKa = 11.84HH540 pKa = 4.99GPGQTMYY547 pKa = 7.93TTQSARR553 pKa = 11.84QEE555 pKa = 3.85LAEE558 pKa = 4.07VVRR561 pKa = 11.84RR562 pKa = 11.84IRR564 pKa = 11.84VKK566 pKa = 11.01GLLSLLNKK574 pKa = 9.72GQEE577 pKa = 4.37LIEE580 pKa = 4.2KK581 pKa = 9.97RR582 pKa = 11.84NTAFGDD588 pKa = 3.62YY589 pKa = 10.39LRR591 pKa = 11.84SLKK594 pKa = 10.82DD595 pKa = 3.32CGLPPTAFSLIEE607 pKa = 4.54SIPVFQVLDD616 pKa = 3.98RR617 pKa = 11.84NQDD620 pKa = 3.91LIKK623 pKa = 10.6QAVIDD628 pKa = 3.97LEE630 pKa = 4.54GTNEE634 pKa = 3.91EE635 pKa = 4.49YY636 pKa = 10.96ALSLDD641 pKa = 3.6EE642 pKa = 4.68SLSIGADD649 pKa = 3.2LVVPDD654 pKa = 5.03PDD656 pKa = 4.15YY657 pKa = 11.65IMSEE661 pKa = 3.92RR662 pKa = 11.84KK663 pKa = 9.1SRR665 pKa = 11.84LIIKK669 pKa = 10.05QNAKK673 pKa = 8.54TLATLLCAFEE683 pKa = 5.35DD684 pKa = 3.98PLIIIDD690 pKa = 6.08DD691 pKa = 4.7DD692 pKa = 4.46DD693 pKa = 4.62QDD695 pKa = 3.81TCIEE699 pKa = 4.48DD700 pKa = 5.15DD701 pKa = 4.28EE702 pKa = 6.42DD703 pKa = 4.56SEE705 pKa = 4.93PDD707 pKa = 3.44SLFQGLYY714 pKa = 10.49DD715 pKa = 5.45EE716 pKa = 5.11EE717 pKa = 5.47PDD719 pKa = 3.29WSAMM723 pKa = 3.64
Molecular weight: 81.97 kDa
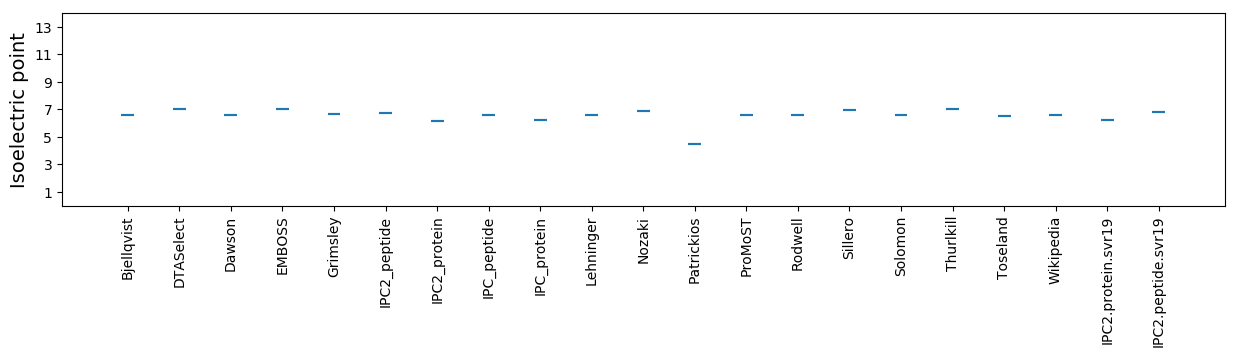
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I5ARE7|A0A2I5ARE7_9VIRU RNA-dependent RNA polymerase OS=Erysiphe necator mitovirus 1 OX=2052561 PE=4 SV=1
MM1 pKa = 7.74AGEE4 pKa = 4.71SCCSVQKK11 pKa = 11.02DD12 pKa = 3.41HH13 pKa = 7.36VLYY16 pKa = 10.18LWGWIEE22 pKa = 3.93TSLKK26 pKa = 10.71HH27 pKa = 6.38KK28 pKa = 10.32GLSHH32 pKa = 7.34TILSLKK38 pKa = 8.14TMRR41 pKa = 11.84NCVTKK46 pKa = 10.27YY47 pKa = 10.13IAGEE51 pKa = 3.79PVKK54 pKa = 10.73VVEE57 pKa = 4.86IRR59 pKa = 11.84LSLTKK64 pKa = 10.61DD65 pKa = 4.32GIPKK69 pKa = 10.42ALGPLVPIIRR79 pKa = 11.84KK80 pKa = 9.46LEE82 pKa = 4.2PQDD85 pKa = 3.54LRR87 pKa = 11.84IVLTLLSVGKK97 pKa = 9.96LYY99 pKa = 10.51PGDD102 pKa = 3.45GTVNTKK108 pKa = 10.66SITDD112 pKa = 3.8PFSGSMSKK120 pKa = 10.04LHH122 pKa = 6.76EE123 pKa = 4.09YY124 pKa = 10.66ALRR127 pKa = 11.84GSFILEE133 pKa = 3.69NLFKK137 pKa = 11.12NQLTQLDD144 pKa = 4.41IEE146 pKa = 4.9QSSNWNAPHH155 pKa = 7.46LSTKK159 pKa = 10.06KK160 pKa = 10.8GPIGHH165 pKa = 7.4ALGTSMIEE173 pKa = 4.6LGHH176 pKa = 7.45LPDD179 pKa = 4.63KK180 pKa = 11.1LLVQIIKK187 pKa = 10.75LGGLKK192 pKa = 10.06LRR194 pKa = 11.84NYY196 pKa = 9.08ILSIIKK202 pKa = 8.66MARR205 pKa = 11.84TISKK209 pKa = 10.28FEE211 pKa = 4.06NSKK214 pKa = 10.88KK215 pKa = 10.18PIPKK219 pKa = 9.64VYY221 pKa = 10.19RR222 pKa = 11.84KK223 pKa = 10.24LSIVKK228 pKa = 10.33DD229 pKa = 3.63KK230 pKa = 10.65EE231 pKa = 4.26LKK233 pKa = 9.98NRR235 pKa = 11.84PIAIFDD241 pKa = 3.62YY242 pKa = 10.11WSQSALKK249 pKa = 8.77PLHH252 pKa = 6.78DD253 pKa = 3.91SVMNLLRR260 pKa = 11.84TFKK263 pKa = 10.62TDD265 pKa = 3.08LTFNQEE271 pKa = 3.73GCEE274 pKa = 4.22EE275 pKa = 4.24IPSLIGPQAKK285 pKa = 9.98LFSLDD290 pKa = 3.65LSSATDD296 pKa = 3.69RR297 pKa = 11.84FPVEE301 pKa = 3.75FQEE304 pKa = 4.79IIVSRR309 pKa = 11.84LIGQEE314 pKa = 3.62RR315 pKa = 11.84AEE317 pKa = 3.84AWRR320 pKa = 11.84YY321 pKa = 10.33IMTEE325 pKa = 4.06FPFNTPEE332 pKa = 4.49GDD334 pKa = 3.61SVKK337 pKa = 10.75YY338 pKa = 8.79NTGQPMGGYY347 pKa = 9.81SSWCIFTLCHH357 pKa = 6.18HH358 pKa = 6.86LVVQIAYY365 pKa = 10.26NEE367 pKa = 4.15AYY369 pKa = 9.23NRR371 pKa = 11.84NWSRR375 pKa = 11.84GARR378 pKa = 11.84SGLPITPKK386 pKa = 10.42LFEE389 pKa = 5.29DD390 pKa = 3.97YY391 pKa = 10.53RR392 pKa = 11.84ILGDD396 pKa = 6.0DD397 pKa = 3.21IVIWDD402 pKa = 3.62EE403 pKa = 3.96MVARR407 pKa = 11.84SYY409 pKa = 11.31KK410 pKa = 10.7SIMADD415 pKa = 3.15LGVEE419 pKa = 4.0ISPFKK424 pKa = 10.31SHH426 pKa = 6.21EE427 pKa = 4.36SYY429 pKa = 8.76HH430 pKa = 4.93TFEE433 pKa = 3.81MAKK436 pKa = 9.65RR437 pKa = 11.84WFHH440 pKa = 6.73KK441 pKa = 9.62GTEE444 pKa = 4.02ITGFPLNNLLEE455 pKa = 4.68AKK457 pKa = 10.08SNPWGIGKK465 pKa = 9.7ALSDD469 pKa = 4.57GIWRR473 pKa = 11.84GWITKK478 pKa = 9.91SNQGVPGNILIRR490 pKa = 11.84RR491 pKa = 11.84LLYY494 pKa = 10.58SLGWSKK500 pKa = 11.0RR501 pKa = 11.84SINNRR506 pKa = 11.84EE507 pKa = 3.62EE508 pKa = 4.1SVRR511 pKa = 11.84DD512 pKa = 3.29SFLFFNFSVTRR523 pKa = 11.84KK524 pKa = 7.52TDD526 pKa = 3.03WVGEE530 pKa = 3.92RR531 pKa = 11.84SVRR534 pKa = 11.84EE535 pKa = 3.82FVSRR539 pKa = 11.84HH540 pKa = 4.99GPGQTMYY547 pKa = 7.93TTQSARR553 pKa = 11.84QEE555 pKa = 3.85LAEE558 pKa = 4.07VVRR561 pKa = 11.84RR562 pKa = 11.84IRR564 pKa = 11.84VKK566 pKa = 11.01GLLSLLNKK574 pKa = 9.72GQEE577 pKa = 4.37LIEE580 pKa = 4.2KK581 pKa = 9.97RR582 pKa = 11.84NTAFGDD588 pKa = 3.62YY589 pKa = 10.39LRR591 pKa = 11.84SLKK594 pKa = 10.82DD595 pKa = 3.32CGLPPTAFSLIEE607 pKa = 4.54SIPVFQVLDD616 pKa = 3.98RR617 pKa = 11.84NQDD620 pKa = 3.91LIKK623 pKa = 10.6QAVIDD628 pKa = 3.97LEE630 pKa = 4.54GTNEE634 pKa = 3.91EE635 pKa = 4.49YY636 pKa = 10.96ALSLDD641 pKa = 3.6EE642 pKa = 4.68SLSIGADD649 pKa = 3.2LVVPDD654 pKa = 5.03PDD656 pKa = 4.15YY657 pKa = 11.65IMSEE661 pKa = 3.92RR662 pKa = 11.84KK663 pKa = 9.1SRR665 pKa = 11.84LIIKK669 pKa = 10.05QNAKK673 pKa = 8.54TLATLLCAFEE683 pKa = 5.35DD684 pKa = 3.98PLIIIDD690 pKa = 6.08DD691 pKa = 4.7DD692 pKa = 4.46DD693 pKa = 4.62QDD695 pKa = 3.81TCIEE699 pKa = 4.48DD700 pKa = 5.15DD701 pKa = 4.28EE702 pKa = 6.42DD703 pKa = 4.56SEE705 pKa = 4.93PDD707 pKa = 3.44SLFQGLYY714 pKa = 10.49DD715 pKa = 5.45EE716 pKa = 5.11EE717 pKa = 5.47PDD719 pKa = 3.29WSAMM723 pKa = 3.64
MM1 pKa = 7.74AGEE4 pKa = 4.71SCCSVQKK11 pKa = 11.02DD12 pKa = 3.41HH13 pKa = 7.36VLYY16 pKa = 10.18LWGWIEE22 pKa = 3.93TSLKK26 pKa = 10.71HH27 pKa = 6.38KK28 pKa = 10.32GLSHH32 pKa = 7.34TILSLKK38 pKa = 8.14TMRR41 pKa = 11.84NCVTKK46 pKa = 10.27YY47 pKa = 10.13IAGEE51 pKa = 3.79PVKK54 pKa = 10.73VVEE57 pKa = 4.86IRR59 pKa = 11.84LSLTKK64 pKa = 10.61DD65 pKa = 4.32GIPKK69 pKa = 10.42ALGPLVPIIRR79 pKa = 11.84KK80 pKa = 9.46LEE82 pKa = 4.2PQDD85 pKa = 3.54LRR87 pKa = 11.84IVLTLLSVGKK97 pKa = 9.96LYY99 pKa = 10.51PGDD102 pKa = 3.45GTVNTKK108 pKa = 10.66SITDD112 pKa = 3.8PFSGSMSKK120 pKa = 10.04LHH122 pKa = 6.76EE123 pKa = 4.09YY124 pKa = 10.66ALRR127 pKa = 11.84GSFILEE133 pKa = 3.69NLFKK137 pKa = 11.12NQLTQLDD144 pKa = 4.41IEE146 pKa = 4.9QSSNWNAPHH155 pKa = 7.46LSTKK159 pKa = 10.06KK160 pKa = 10.8GPIGHH165 pKa = 7.4ALGTSMIEE173 pKa = 4.6LGHH176 pKa = 7.45LPDD179 pKa = 4.63KK180 pKa = 11.1LLVQIIKK187 pKa = 10.75LGGLKK192 pKa = 10.06LRR194 pKa = 11.84NYY196 pKa = 9.08ILSIIKK202 pKa = 8.66MARR205 pKa = 11.84TISKK209 pKa = 10.28FEE211 pKa = 4.06NSKK214 pKa = 10.88KK215 pKa = 10.18PIPKK219 pKa = 9.64VYY221 pKa = 10.19RR222 pKa = 11.84KK223 pKa = 10.24LSIVKK228 pKa = 10.33DD229 pKa = 3.63KK230 pKa = 10.65EE231 pKa = 4.26LKK233 pKa = 9.98NRR235 pKa = 11.84PIAIFDD241 pKa = 3.62YY242 pKa = 10.11WSQSALKK249 pKa = 8.77PLHH252 pKa = 6.78DD253 pKa = 3.91SVMNLLRR260 pKa = 11.84TFKK263 pKa = 10.62TDD265 pKa = 3.08LTFNQEE271 pKa = 3.73GCEE274 pKa = 4.22EE275 pKa = 4.24IPSLIGPQAKK285 pKa = 9.98LFSLDD290 pKa = 3.65LSSATDD296 pKa = 3.69RR297 pKa = 11.84FPVEE301 pKa = 3.75FQEE304 pKa = 4.79IIVSRR309 pKa = 11.84LIGQEE314 pKa = 3.62RR315 pKa = 11.84AEE317 pKa = 3.84AWRR320 pKa = 11.84YY321 pKa = 10.33IMTEE325 pKa = 4.06FPFNTPEE332 pKa = 4.49GDD334 pKa = 3.61SVKK337 pKa = 10.75YY338 pKa = 8.79NTGQPMGGYY347 pKa = 9.81SSWCIFTLCHH357 pKa = 6.18HH358 pKa = 6.86LVVQIAYY365 pKa = 10.26NEE367 pKa = 4.15AYY369 pKa = 9.23NRR371 pKa = 11.84NWSRR375 pKa = 11.84GARR378 pKa = 11.84SGLPITPKK386 pKa = 10.42LFEE389 pKa = 5.29DD390 pKa = 3.97YY391 pKa = 10.53RR392 pKa = 11.84ILGDD396 pKa = 6.0DD397 pKa = 3.21IVIWDD402 pKa = 3.62EE403 pKa = 3.96MVARR407 pKa = 11.84SYY409 pKa = 11.31KK410 pKa = 10.7SIMADD415 pKa = 3.15LGVEE419 pKa = 4.0ISPFKK424 pKa = 10.31SHH426 pKa = 6.21EE427 pKa = 4.36SYY429 pKa = 8.76HH430 pKa = 4.93TFEE433 pKa = 3.81MAKK436 pKa = 9.65RR437 pKa = 11.84WFHH440 pKa = 6.73KK441 pKa = 9.62GTEE444 pKa = 4.02ITGFPLNNLLEE455 pKa = 4.68AKK457 pKa = 10.08SNPWGIGKK465 pKa = 9.7ALSDD469 pKa = 4.57GIWRR473 pKa = 11.84GWITKK478 pKa = 9.91SNQGVPGNILIRR490 pKa = 11.84RR491 pKa = 11.84LLYY494 pKa = 10.58SLGWSKK500 pKa = 11.0RR501 pKa = 11.84SINNRR506 pKa = 11.84EE507 pKa = 3.62EE508 pKa = 4.1SVRR511 pKa = 11.84DD512 pKa = 3.29SFLFFNFSVTRR523 pKa = 11.84KK524 pKa = 7.52TDD526 pKa = 3.03WVGEE530 pKa = 3.92RR531 pKa = 11.84SVRR534 pKa = 11.84EE535 pKa = 3.82FVSRR539 pKa = 11.84HH540 pKa = 4.99GPGQTMYY547 pKa = 7.93TTQSARR553 pKa = 11.84QEE555 pKa = 3.85LAEE558 pKa = 4.07VVRR561 pKa = 11.84RR562 pKa = 11.84IRR564 pKa = 11.84VKK566 pKa = 11.01GLLSLLNKK574 pKa = 9.72GQEE577 pKa = 4.37LIEE580 pKa = 4.2KK581 pKa = 9.97RR582 pKa = 11.84NTAFGDD588 pKa = 3.62YY589 pKa = 10.39LRR591 pKa = 11.84SLKK594 pKa = 10.82DD595 pKa = 3.32CGLPPTAFSLIEE607 pKa = 4.54SIPVFQVLDD616 pKa = 3.98RR617 pKa = 11.84NQDD620 pKa = 3.91LIKK623 pKa = 10.6QAVIDD628 pKa = 3.97LEE630 pKa = 4.54GTNEE634 pKa = 3.91EE635 pKa = 4.49YY636 pKa = 10.96ALSLDD641 pKa = 3.6EE642 pKa = 4.68SLSIGADD649 pKa = 3.2LVVPDD654 pKa = 5.03PDD656 pKa = 4.15YY657 pKa = 11.65IMSEE661 pKa = 3.92RR662 pKa = 11.84KK663 pKa = 9.1SRR665 pKa = 11.84LIIKK669 pKa = 10.05QNAKK673 pKa = 8.54TLATLLCAFEE683 pKa = 5.35DD684 pKa = 3.98PLIIIDD690 pKa = 6.08DD691 pKa = 4.7DD692 pKa = 4.46DD693 pKa = 4.62QDD695 pKa = 3.81TCIEE699 pKa = 4.48DD700 pKa = 5.15DD701 pKa = 4.28EE702 pKa = 6.42DD703 pKa = 4.56SEE705 pKa = 4.93PDD707 pKa = 3.44SLFQGLYY714 pKa = 10.49DD715 pKa = 5.45EE716 pKa = 5.11EE717 pKa = 5.47PDD719 pKa = 3.29WSAMM723 pKa = 3.64
Molecular weight: 81.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
723 |
723 |
723 |
723.0 |
81.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.426 ± 0.0 | 1.245 ± 0.0 |
5.947 ± 0.0 | 6.639 ± 0.0 |
3.873 ± 0.0 | 6.501 ± 0.0 |
1.936 ± 0.0 | 8.022 ± 0.0 |
6.777 ± 0.0 | 11.48 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.936 ± 0.0 | 4.011 ± 0.0 |
4.841 ± 0.0 | 3.32 ± 0.0 |
5.394 ± 0.0 | 8.575 ± 0.0 |
5.118 ± 0.0 | 4.979 ± 0.0 |
2.075 ± 0.0 | 2.905 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
