
Cellulomonas hominis
Taxonomy:
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
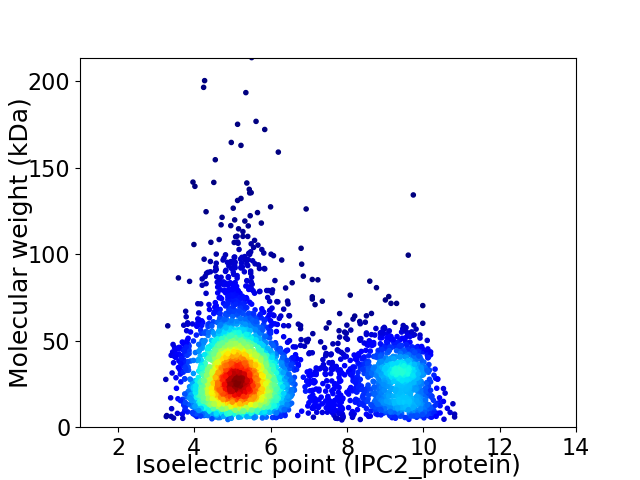
Virtual 2D-PAGE plot for 4041 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A511F851|A0A511F851_9CELL DNA-binding transcriptional MerR regulator OS=Cellulomonas hominis OX=156981 GN=CHO01_05010 PE=4 SV=1
MM1 pKa = 7.57PAGPAVAAPADD12 pKa = 3.99DD13 pKa = 4.24CAAPTRR19 pKa = 11.84TVTGSTSGAPLDD31 pKa = 3.82VAAGEE36 pKa = 4.44VVLIAGGTQSGGVNSLPAGATLCVAAGATLAPAYY70 pKa = 10.21VNNAAGTVYY79 pKa = 10.3VAPGGTASLPSIATNAGFVLDD100 pKa = 4.03NAGTTTVAGLNVNGPGTVRR119 pKa = 11.84NAAGATLAVTGQLAPAAGTVVNDD142 pKa = 3.9GTLALPGGATVNGNVSLVNNGTLTAGGTTTISGAFRR178 pKa = 11.84NAGTATLGGDD188 pKa = 3.59VIVNGGGSLHH198 pKa = 6.16NLCIIDD204 pKa = 3.8AAGGLTTNAPGSTNTGLVALGGGLQVSGSGSWSQTPTGATTATTLTDD251 pKa = 3.84DD252 pKa = 4.52GAVTGYY258 pKa = 10.57GRR260 pKa = 11.84YY261 pKa = 9.98AFTGTTSVQGTFAGDD276 pKa = 3.36LALVPIFLDD285 pKa = 3.65VPGTTFPYY293 pKa = 10.02FDD295 pKa = 3.7VVNGTVANVDD305 pKa = 3.52RR306 pKa = 11.84RR307 pKa = 11.84DD308 pKa = 3.58LTIPGPADD316 pKa = 3.62YY317 pKa = 8.01PAPDD321 pKa = 4.47CADD324 pKa = 3.64PAAPPSADD332 pKa = 3.28VQVAKK337 pKa = 9.99TGPASVLPDD346 pKa = 3.22GTLTYY351 pKa = 10.42TVTVTNAGPDD361 pKa = 3.28AAEE364 pKa = 4.64DD365 pKa = 3.73VTVTDD370 pKa = 4.11TLPPTLLDD378 pKa = 3.77PAADD382 pKa = 3.51AGGVVAGGTVTWSLGTLAAGQTVTLTASGRR412 pKa = 11.84APADD416 pKa = 3.34GTLVNVVSATSTTPDD431 pKa = 3.63PDD433 pKa = 3.23ASNNDD438 pKa = 2.82GSADD442 pKa = 3.88SEE444 pKa = 4.82TVTTDD449 pKa = 3.34VEE451 pKa = 4.36PSPPDD456 pKa = 3.38TGPPTAAPLTRR467 pKa = 11.84DD468 pKa = 4.08GPPGLPIVGVLDD480 pKa = 4.43GSSPDD485 pKa = 3.61PDD487 pKa = 3.32LQLRR491 pKa = 11.84YY492 pKa = 10.03SVVDD496 pKa = 4.07GPTSGRR502 pKa = 11.84VIALPNGLFGYY513 pKa = 10.4LPDD516 pKa = 5.26DD517 pKa = 4.24GFVGTDD523 pKa = 2.74TFTYY527 pKa = 9.86RR528 pKa = 11.84VCDD531 pKa = 3.44NQTPAEE537 pKa = 4.3CSAPATVTLIIHH549 pKa = 6.33PRR551 pKa = 11.84AVNDD555 pKa = 3.4EE556 pKa = 4.37TTTLMDD562 pKa = 3.3QPVTIDD568 pKa = 3.58VLANDD573 pKa = 4.24PGGAVLSSNLDD584 pKa = 3.16ASASHH589 pKa = 6.9GLTQIDD595 pKa = 4.53PATGTVTYY603 pKa = 9.7TPGPGYY609 pKa = 10.62LGDD612 pKa = 3.56DD613 pKa = 3.6TFVYY617 pKa = 9.91RR618 pKa = 11.84ACAPADD624 pKa = 4.07PGDD627 pKa = 3.86CALAVVVVHH636 pKa = 5.99VVPDD640 pKa = 4.03NLPPLAPPLAMEE652 pKa = 4.46TTVGAPVTGAPAVSDD667 pKa = 3.96PDD669 pKa = 3.64GDD671 pKa = 4.09AVALASVFPPTTGDD685 pKa = 3.07ATATTTTTTYY695 pKa = 9.94TPPPGFAGRR704 pKa = 11.84AVYY707 pKa = 10.09QYY709 pKa = 7.55TVCDD713 pKa = 3.87DD714 pKa = 4.1GVPVLCATGLVTVLVDD730 pKa = 4.1PVAGDD735 pKa = 4.24DD736 pKa = 4.63SATTPAGTPVTVDD749 pKa = 2.91VAANDD754 pKa = 4.08LGTVLPPDD762 pKa = 3.85VATPPANGTVALAGGALVYY781 pKa = 10.12TPAPGFVGTDD791 pKa = 2.81TFTYY795 pKa = 9.78RR796 pKa = 11.84ICADD800 pKa = 4.1DD801 pKa = 4.12VSPACDD807 pKa = 2.98TATVTVVVTGDD818 pKa = 4.11GPPVDD823 pKa = 4.98PGDD826 pKa = 4.52PGDD829 pKa = 4.34GGAGGTGGGSGTGAAPGGGGLPVTGADD856 pKa = 3.74AAPLAALAAALLAAGAAALTARR878 pKa = 11.84RR879 pKa = 11.84RR880 pKa = 11.84ATAPARR886 pKa = 11.84ARR888 pKa = 11.84RR889 pKa = 11.84PGPRR893 pKa = 11.84RR894 pKa = 3.3
MM1 pKa = 7.57PAGPAVAAPADD12 pKa = 3.99DD13 pKa = 4.24CAAPTRR19 pKa = 11.84TVTGSTSGAPLDD31 pKa = 3.82VAAGEE36 pKa = 4.44VVLIAGGTQSGGVNSLPAGATLCVAAGATLAPAYY70 pKa = 10.21VNNAAGTVYY79 pKa = 10.3VAPGGTASLPSIATNAGFVLDD100 pKa = 4.03NAGTTTVAGLNVNGPGTVRR119 pKa = 11.84NAAGATLAVTGQLAPAAGTVVNDD142 pKa = 3.9GTLALPGGATVNGNVSLVNNGTLTAGGTTTISGAFRR178 pKa = 11.84NAGTATLGGDD188 pKa = 3.59VIVNGGGSLHH198 pKa = 6.16NLCIIDD204 pKa = 3.8AAGGLTTNAPGSTNTGLVALGGGLQVSGSGSWSQTPTGATTATTLTDD251 pKa = 3.84DD252 pKa = 4.52GAVTGYY258 pKa = 10.57GRR260 pKa = 11.84YY261 pKa = 9.98AFTGTTSVQGTFAGDD276 pKa = 3.36LALVPIFLDD285 pKa = 3.65VPGTTFPYY293 pKa = 10.02FDD295 pKa = 3.7VVNGTVANVDD305 pKa = 3.52RR306 pKa = 11.84RR307 pKa = 11.84DD308 pKa = 3.58LTIPGPADD316 pKa = 3.62YY317 pKa = 8.01PAPDD321 pKa = 4.47CADD324 pKa = 3.64PAAPPSADD332 pKa = 3.28VQVAKK337 pKa = 9.99TGPASVLPDD346 pKa = 3.22GTLTYY351 pKa = 10.42TVTVTNAGPDD361 pKa = 3.28AAEE364 pKa = 4.64DD365 pKa = 3.73VTVTDD370 pKa = 4.11TLPPTLLDD378 pKa = 3.77PAADD382 pKa = 3.51AGGVVAGGTVTWSLGTLAAGQTVTLTASGRR412 pKa = 11.84APADD416 pKa = 3.34GTLVNVVSATSTTPDD431 pKa = 3.63PDD433 pKa = 3.23ASNNDD438 pKa = 2.82GSADD442 pKa = 3.88SEE444 pKa = 4.82TVTTDD449 pKa = 3.34VEE451 pKa = 4.36PSPPDD456 pKa = 3.38TGPPTAAPLTRR467 pKa = 11.84DD468 pKa = 4.08GPPGLPIVGVLDD480 pKa = 4.43GSSPDD485 pKa = 3.61PDD487 pKa = 3.32LQLRR491 pKa = 11.84YY492 pKa = 10.03SVVDD496 pKa = 4.07GPTSGRR502 pKa = 11.84VIALPNGLFGYY513 pKa = 10.4LPDD516 pKa = 5.26DD517 pKa = 4.24GFVGTDD523 pKa = 2.74TFTYY527 pKa = 9.86RR528 pKa = 11.84VCDD531 pKa = 3.44NQTPAEE537 pKa = 4.3CSAPATVTLIIHH549 pKa = 6.33PRR551 pKa = 11.84AVNDD555 pKa = 3.4EE556 pKa = 4.37TTTLMDD562 pKa = 3.3QPVTIDD568 pKa = 3.58VLANDD573 pKa = 4.24PGGAVLSSNLDD584 pKa = 3.16ASASHH589 pKa = 6.9GLTQIDD595 pKa = 4.53PATGTVTYY603 pKa = 9.7TPGPGYY609 pKa = 10.62LGDD612 pKa = 3.56DD613 pKa = 3.6TFVYY617 pKa = 9.91RR618 pKa = 11.84ACAPADD624 pKa = 4.07PGDD627 pKa = 3.86CALAVVVVHH636 pKa = 5.99VVPDD640 pKa = 4.03NLPPLAPPLAMEE652 pKa = 4.46TTVGAPVTGAPAVSDD667 pKa = 3.96PDD669 pKa = 3.64GDD671 pKa = 4.09AVALASVFPPTTGDD685 pKa = 3.07ATATTTTTTYY695 pKa = 9.94TPPPGFAGRR704 pKa = 11.84AVYY707 pKa = 10.09QYY709 pKa = 7.55TVCDD713 pKa = 3.87DD714 pKa = 4.1GVPVLCATGLVTVLVDD730 pKa = 4.1PVAGDD735 pKa = 4.24DD736 pKa = 4.63SATTPAGTPVTVDD749 pKa = 2.91VAANDD754 pKa = 4.08LGTVLPPDD762 pKa = 3.85VATPPANGTVALAGGALVYY781 pKa = 10.12TPAPGFVGTDD791 pKa = 2.81TFTYY795 pKa = 9.78RR796 pKa = 11.84ICADD800 pKa = 4.1DD801 pKa = 4.12VSPACDD807 pKa = 2.98TATVTVVVTGDD818 pKa = 4.11GPPVDD823 pKa = 4.98PGDD826 pKa = 4.52PGDD829 pKa = 4.34GGAGGTGGGSGTGAAPGGGGLPVTGADD856 pKa = 3.74AAPLAALAAALLAAGAAALTARR878 pKa = 11.84RR879 pKa = 11.84RR880 pKa = 11.84ATAPARR886 pKa = 11.84ARR888 pKa = 11.84RR889 pKa = 11.84PGPRR893 pKa = 11.84RR894 pKa = 3.3
Molecular weight: 86.31 kDa
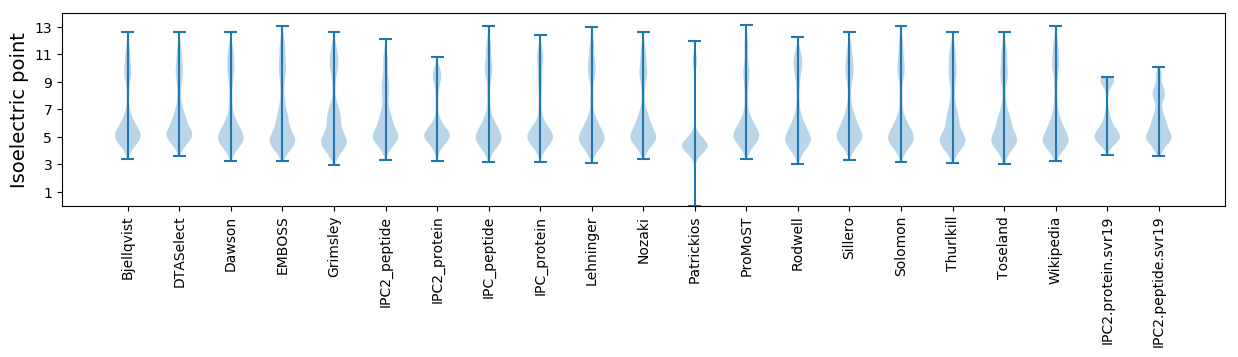
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A511F8J5|A0A511F8J5_9CELL AcrR family transcriptional regulator OS=Cellulomonas hominis OX=156981 GN=CHO01_06880 PE=4 SV=1
MM1 pKa = 7.39GRR3 pKa = 11.84RR4 pKa = 11.84GSVRR8 pKa = 11.84LWGGVAFAALVLVAVGWAFAPAITLLVGVVVAALAVPVGIVLVATAALSRR58 pKa = 11.84RR59 pKa = 3.97
MM1 pKa = 7.39GRR3 pKa = 11.84RR4 pKa = 11.84GSVRR8 pKa = 11.84LWGGVAFAALVLVAVGWAFAPAITLLVGVVVAALAVPVGIVLVATAALSRR58 pKa = 11.84RR59 pKa = 3.97
Molecular weight: 5.91 kDa
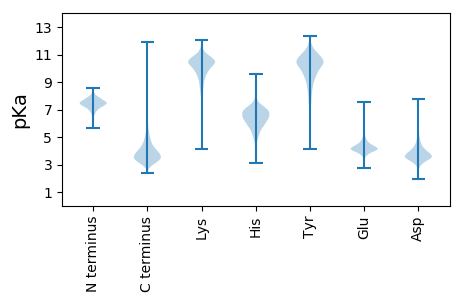
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1292075 |
39 |
2005 |
319.7 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.344 ± 0.068 | 0.559 ± 0.009 |
6.441 ± 0.036 | 5.19 ± 0.036 |
2.434 ± 0.022 | 9.555 ± 0.037 |
2.019 ± 0.017 | 2.766 ± 0.029 |
1.277 ± 0.023 | 10.415 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.574 ± 0.014 | 1.383 ± 0.016 |
6.213 ± 0.033 | 2.614 ± 0.021 |
8.1 ± 0.044 | 4.591 ± 0.031 |
6.121 ± 0.044 | 10.043 ± 0.04 |
1.556 ± 0.018 | 1.805 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
