
Eidolon helvum papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoupsilonpapillomavirus; Dyoupsilonpapillomavirus 1
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
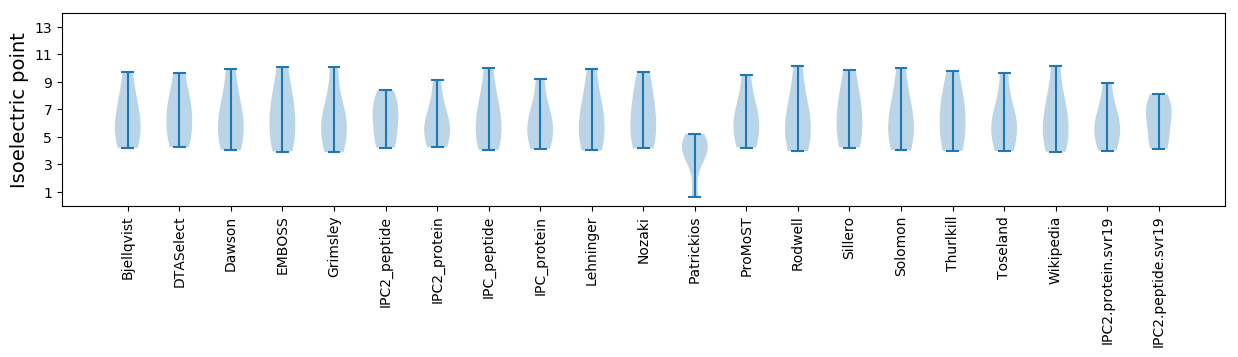
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4TH86|S4TH86_9PAPI Protein E7 OS=Eidolon helvum papillomavirus 1 OX=1163701 GN=E7 PE=3 SV=1
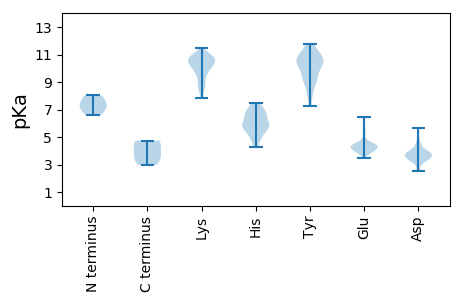
MM1 pKa = 7.8RR2 pKa = 11.84IEE4 pKa = 4.63LPSLQEE10 pKa = 4.25LVSQEE15 pKa = 3.81NPEE18 pKa = 4.39AVDD21 pKa = 3.91LNCYY25 pKa = 10.29EE26 pKa = 4.24IMSSDD31 pKa = 3.56EE32 pKa = 4.26EE33 pKa = 4.37EE34 pKa = 4.28VDD36 pKa = 4.15EE37 pKa = 4.65EE38 pKa = 4.45LPLYY42 pKa = 10.51SVAVHH47 pKa = 6.58CGSCRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84VRR56 pKa = 11.84LFCAASRR63 pKa = 11.84EE64 pKa = 4.17ALHH67 pKa = 7.04LLEE70 pKa = 5.54CLLFEE75 pKa = 5.53DD76 pKa = 5.64LLAFLCPGCALAGEE90 pKa = 4.86DD91 pKa = 4.44GGG93 pKa = 4.68
MM1 pKa = 7.8RR2 pKa = 11.84IEE4 pKa = 4.63LPSLQEE10 pKa = 4.25LVSQEE15 pKa = 3.81NPEE18 pKa = 4.39AVDD21 pKa = 3.91LNCYY25 pKa = 10.29EE26 pKa = 4.24IMSSDD31 pKa = 3.56EE32 pKa = 4.26EE33 pKa = 4.37EE34 pKa = 4.28VDD36 pKa = 4.15EE37 pKa = 4.65EE38 pKa = 4.45LPLYY42 pKa = 10.51SVAVHH47 pKa = 6.58CGSCRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84VRR56 pKa = 11.84LFCAASRR63 pKa = 11.84EE64 pKa = 4.17ALHH67 pKa = 7.04LLEE70 pKa = 5.54CLLFEE75 pKa = 5.53DD76 pKa = 5.64LLAFLCPGCALAGEE90 pKa = 4.86DD91 pKa = 4.44GGG93 pKa = 4.68
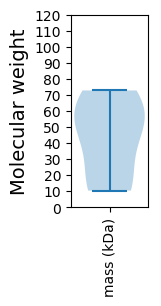
Molecular weight: 10.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S4TH17|S4TH17_9PAPI Protein E6 OS=Eidolon helvum papillomavirus 1 OX=1163701 GN=E6 PE=3 SV=1
MM1 pKa = 6.96EE2 pKa = 4.5TEE4 pKa = 4.1KK5 pKa = 10.83LAKK8 pKa = 9.83RR9 pKa = 11.84LSSLEE14 pKa = 3.95EE15 pKa = 4.21EE16 pKa = 4.45ILRR19 pKa = 11.84HH20 pKa = 4.99YY21 pKa = 10.71EE22 pKa = 3.83KK23 pKa = 10.58DD24 pKa = 3.45SKK26 pKa = 11.01RR27 pKa = 11.84LEE29 pKa = 3.96DD30 pKa = 5.64QIRR33 pKa = 11.84FWNLMRR39 pKa = 11.84RR40 pKa = 11.84EE41 pKa = 3.89QALMHH46 pKa = 6.85FIRR49 pKa = 11.84KK50 pKa = 8.93QGVHH54 pKa = 5.5RR55 pKa = 11.84VGLRR59 pKa = 11.84PIPSLMASEE68 pKa = 5.32AEE70 pKa = 3.88AKK72 pKa = 10.17KK73 pKa = 10.28AIEE76 pKa = 4.11MEE78 pKa = 4.37VVLTSLSKK86 pKa = 10.67SQFAFEE92 pKa = 4.25SWGLRR97 pKa = 11.84DD98 pKa = 4.12TSWEE102 pKa = 4.11LYY104 pKa = 9.42KK105 pKa = 10.86APPEE109 pKa = 4.12NTLKK113 pKa = 10.83KK114 pKa = 10.4YY115 pKa = 9.02GQQVEE120 pKa = 4.45LTYY123 pKa = 11.42DD124 pKa = 3.56NDD126 pKa = 3.88PDD128 pKa = 3.68NRR130 pKa = 11.84SAEE133 pKa = 4.32TLWGGLYY140 pKa = 10.09IMYY143 pKa = 10.51EE144 pKa = 3.74NDD146 pKa = 2.71DD147 pKa = 3.44WRR149 pKa = 11.84RR150 pKa = 11.84VTSAVDD156 pKa = 3.22EE157 pKa = 4.37KK158 pKa = 11.18GIYY161 pKa = 9.63YY162 pKa = 10.06VDD164 pKa = 3.84PDD166 pKa = 3.89GLRR169 pKa = 11.84IYY171 pKa = 10.25YY172 pKa = 10.64VNFEE176 pKa = 4.38ALADD180 pKa = 4.39RR181 pKa = 11.84YY182 pKa = 10.83GSTGAYY188 pKa = 9.34KK189 pKa = 10.92VMAGSDD195 pKa = 4.26LIAVVGSDD203 pKa = 3.55TDD205 pKa = 4.45SEE207 pKa = 4.48DD208 pKa = 3.23TTAPDD213 pKa = 3.82APSIAARR220 pKa = 11.84RR221 pKa = 11.84VSSPQTPKK229 pKa = 10.49PSPKK233 pKa = 8.85KK234 pKa = 8.57TPPRR238 pKa = 11.84KK239 pKa = 7.86PPRR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84VSTPTRR251 pKa = 11.84RR252 pKa = 11.84QRR254 pKa = 11.84RR255 pKa = 11.84GEE257 pKa = 3.87PGSRR261 pKa = 11.84NRR263 pKa = 11.84RR264 pKa = 11.84GARR267 pKa = 11.84ATAGCTRR274 pKa = 11.84GEE276 pKa = 4.38CTPPSPSQVGRR287 pKa = 11.84RR288 pKa = 11.84HH289 pKa = 5.85QSTPPKK295 pKa = 10.28NLGRR299 pKa = 11.84LGRR302 pKa = 11.84LLAEE306 pKa = 5.11AYY308 pKa = 8.96DD309 pKa = 4.18PPILVCRR316 pKa = 11.84GAANTLKK323 pKa = 9.98CLRR326 pKa = 11.84YY327 pKa = 9.22RR328 pKa = 11.84VHH330 pKa = 7.07LKK332 pKa = 10.49HH333 pKa = 6.84SNLYY337 pKa = 9.13DD338 pKa = 3.75TVSTTWNWTKK348 pKa = 10.83GSYY351 pKa = 8.08PQSRR355 pKa = 11.84IIYY358 pKa = 8.36LFNSKK363 pKa = 8.95HH364 pKa = 4.29QRR366 pKa = 11.84EE367 pKa = 4.5SFLEE371 pKa = 4.18HH372 pKa = 5.86VKK374 pKa = 10.88LPASVTHH381 pKa = 5.22YY382 pKa = 9.92TGFLNGII389 pKa = 4.22
MM1 pKa = 6.96EE2 pKa = 4.5TEE4 pKa = 4.1KK5 pKa = 10.83LAKK8 pKa = 9.83RR9 pKa = 11.84LSSLEE14 pKa = 3.95EE15 pKa = 4.21EE16 pKa = 4.45ILRR19 pKa = 11.84HH20 pKa = 4.99YY21 pKa = 10.71EE22 pKa = 3.83KK23 pKa = 10.58DD24 pKa = 3.45SKK26 pKa = 11.01RR27 pKa = 11.84LEE29 pKa = 3.96DD30 pKa = 5.64QIRR33 pKa = 11.84FWNLMRR39 pKa = 11.84RR40 pKa = 11.84EE41 pKa = 3.89QALMHH46 pKa = 6.85FIRR49 pKa = 11.84KK50 pKa = 8.93QGVHH54 pKa = 5.5RR55 pKa = 11.84VGLRR59 pKa = 11.84PIPSLMASEE68 pKa = 5.32AEE70 pKa = 3.88AKK72 pKa = 10.17KK73 pKa = 10.28AIEE76 pKa = 4.11MEE78 pKa = 4.37VVLTSLSKK86 pKa = 10.67SQFAFEE92 pKa = 4.25SWGLRR97 pKa = 11.84DD98 pKa = 4.12TSWEE102 pKa = 4.11LYY104 pKa = 9.42KK105 pKa = 10.86APPEE109 pKa = 4.12NTLKK113 pKa = 10.83KK114 pKa = 10.4YY115 pKa = 9.02GQQVEE120 pKa = 4.45LTYY123 pKa = 11.42DD124 pKa = 3.56NDD126 pKa = 3.88PDD128 pKa = 3.68NRR130 pKa = 11.84SAEE133 pKa = 4.32TLWGGLYY140 pKa = 10.09IMYY143 pKa = 10.51EE144 pKa = 3.74NDD146 pKa = 2.71DD147 pKa = 3.44WRR149 pKa = 11.84RR150 pKa = 11.84VTSAVDD156 pKa = 3.22EE157 pKa = 4.37KK158 pKa = 11.18GIYY161 pKa = 9.63YY162 pKa = 10.06VDD164 pKa = 3.84PDD166 pKa = 3.89GLRR169 pKa = 11.84IYY171 pKa = 10.25YY172 pKa = 10.64VNFEE176 pKa = 4.38ALADD180 pKa = 4.39RR181 pKa = 11.84YY182 pKa = 10.83GSTGAYY188 pKa = 9.34KK189 pKa = 10.92VMAGSDD195 pKa = 4.26LIAVVGSDD203 pKa = 3.55TDD205 pKa = 4.45SEE207 pKa = 4.48DD208 pKa = 3.23TTAPDD213 pKa = 3.82APSIAARR220 pKa = 11.84RR221 pKa = 11.84VSSPQTPKK229 pKa = 10.49PSPKK233 pKa = 8.85KK234 pKa = 8.57TPPRR238 pKa = 11.84KK239 pKa = 7.86PPRR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84VSTPTRR251 pKa = 11.84RR252 pKa = 11.84QRR254 pKa = 11.84RR255 pKa = 11.84GEE257 pKa = 3.87PGSRR261 pKa = 11.84NRR263 pKa = 11.84RR264 pKa = 11.84GARR267 pKa = 11.84ATAGCTRR274 pKa = 11.84GEE276 pKa = 4.38CTPPSPSQVGRR287 pKa = 11.84RR288 pKa = 11.84HH289 pKa = 5.85QSTPPKK295 pKa = 10.28NLGRR299 pKa = 11.84LGRR302 pKa = 11.84LLAEE306 pKa = 5.11AYY308 pKa = 8.96DD309 pKa = 4.18PPILVCRR316 pKa = 11.84GAANTLKK323 pKa = 9.98CLRR326 pKa = 11.84YY327 pKa = 9.22RR328 pKa = 11.84VHH330 pKa = 7.07LKK332 pKa = 10.49HH333 pKa = 6.84SNLYY337 pKa = 9.13DD338 pKa = 3.75TVSTTWNWTKK348 pKa = 10.83GSYY351 pKa = 8.08PQSRR355 pKa = 11.84IIYY358 pKa = 8.36LFNSKK363 pKa = 8.95HH364 pKa = 4.29QRR366 pKa = 11.84EE367 pKa = 4.5SFLEE371 pKa = 4.18HH372 pKa = 5.86VKK374 pKa = 10.88LPASVTHH381 pKa = 5.22YY382 pKa = 9.92TGFLNGII389 pKa = 4.22
Molecular weight: 44.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2311 |
93 |
640 |
385.2 |
43.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.928 ± 0.27 | 1.861 ± 0.691 |
6.145 ± 0.297 | 7.14 ± 0.427 |
4.414 ± 0.591 | 6.621 ± 0.499 |
2.077 ± 0.155 | 4.717 ± 0.768 |
5.019 ± 0.719 | 8.135 ± 0.747 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.688 ± 0.297 | 3.721 ± 0.667 |
6.318 ± 0.884 | 4.37 ± 0.518 |
6.707 ± 0.764 | 6.404 ± 0.634 |
7.572 ± 0.864 | 6.491 ± 0.786 |
1.255 ± 0.33 | 3.418 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
