
Candidatus Regiella insecticola LSR1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Enterobacteriaceae incertae sedis; ant, tsetse, mealybug, aphid, etc. endosymbionts; aphid secondary symbionts; Candidatus Regiella; Candidatus Regiella insecticola
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
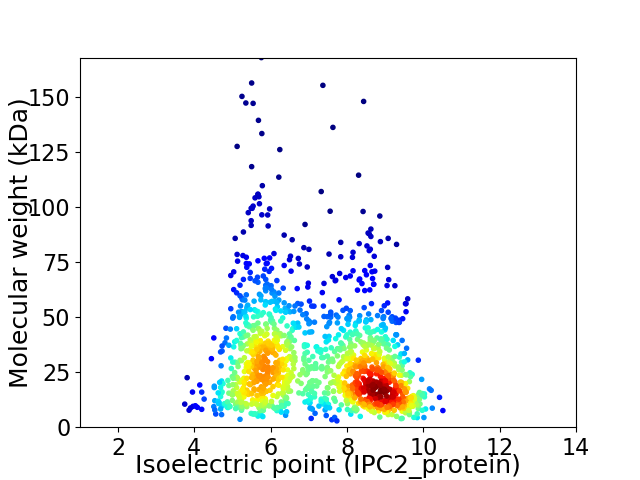
Virtual 2D-PAGE plot for 1368 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0WV44|E0WV44_9ENTR Putative plasmid assoicated protein OS=Candidatus Regiella insecticola LSR1 OX=663321 GN=REG_1959 PE=4 SV=1
MM1 pKa = 7.62FYY3 pKa = 10.64EE4 pKa = 4.02QDD6 pKa = 3.43FDD8 pKa = 4.24QLQAGNDD15 pKa = 3.19NFTWHH20 pKa = 6.07VALSDD25 pKa = 4.59PLPEE29 pKa = 5.38DD30 pKa = 2.86NWSGYY35 pKa = 8.52TGFIHH40 pKa = 6.84NVLLEE45 pKa = 4.48NYY47 pKa = 9.74LKK49 pKa = 10.7DD50 pKa = 3.42HH51 pKa = 7.35PAPEE55 pKa = 4.37DD56 pKa = 3.67CEE58 pKa = 5.73FYY60 pKa = 10.28MCGPPMMNAAVISMLKK76 pKa = 10.09NLGVEE81 pKa = 4.84DD82 pKa = 4.41EE83 pKa = 5.42NIMLDD88 pKa = 4.04DD89 pKa = 4.31FGGG92 pKa = 3.45
MM1 pKa = 7.62FYY3 pKa = 10.64EE4 pKa = 4.02QDD6 pKa = 3.43FDD8 pKa = 4.24QLQAGNDD15 pKa = 3.19NFTWHH20 pKa = 6.07VALSDD25 pKa = 4.59PLPEE29 pKa = 5.38DD30 pKa = 2.86NWSGYY35 pKa = 8.52TGFIHH40 pKa = 6.84NVLLEE45 pKa = 4.48NYY47 pKa = 9.74LKK49 pKa = 10.7DD50 pKa = 3.42HH51 pKa = 7.35PAPEE55 pKa = 4.37DD56 pKa = 3.67CEE58 pKa = 5.73FYY60 pKa = 10.28MCGPPMMNAAVISMLKK76 pKa = 10.09NLGVEE81 pKa = 4.84DD82 pKa = 4.41EE83 pKa = 5.42NIMLDD88 pKa = 4.04DD89 pKa = 4.31FGGG92 pKa = 3.45
Molecular weight: 10.49 kDa
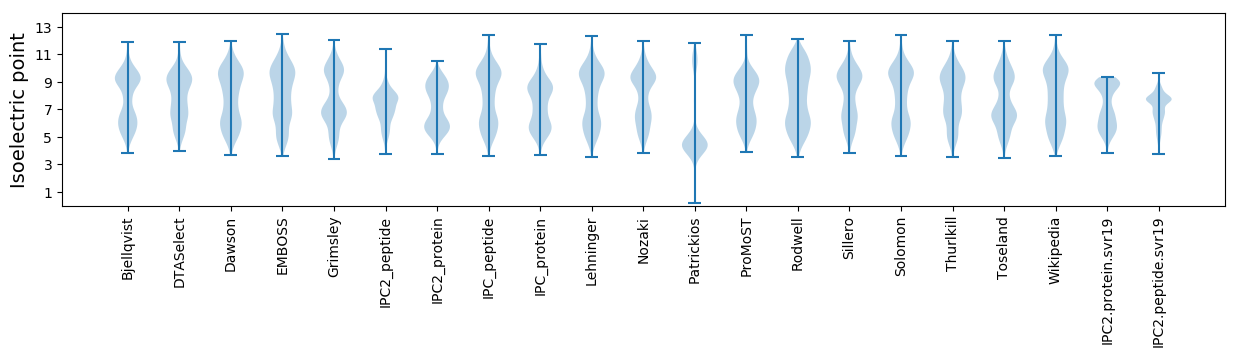
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0WTP1|E0WTP1_9ENTR Uncharacterized protein OS=Candidatus Regiella insecticola LSR1 OX=663321 GN=REG_1411 PE=4 SV=1
MM1 pKa = 7.88LPQCNMKK8 pKa = 10.14SRR10 pKa = 11.84CRR12 pKa = 11.84YY13 pKa = 8.73GFTHH17 pKa = 7.19RR18 pKa = 11.84RR19 pKa = 11.84LQHH22 pKa = 6.15LPCGGKK28 pKa = 7.6TLILCFRR35 pKa = 11.84LQRR38 pKa = 11.84FCCTNSRR45 pKa = 11.84CPQRR49 pKa = 11.84TFQEE53 pKa = 4.68SISTLAPRR61 pKa = 11.84WQRR64 pKa = 11.84SSRR67 pKa = 11.84QLVEE71 pKa = 3.6MTTRR75 pKa = 11.84LAYY78 pKa = 10.42SFGVRR83 pKa = 11.84VSGDD87 pKa = 3.24TLIRR91 pKa = 11.84RR92 pKa = 11.84IMRR95 pKa = 11.84LPLSEE100 pKa = 4.78PEE102 pKa = 4.07QLTHH106 pKa = 7.61IGVDD110 pKa = 3.06DD111 pKa = 3.85WAWRR115 pKa = 11.84KK116 pKa = 8.0GHH118 pKa = 6.8HH119 pKa = 6.54YY120 pKa = 9.44GTIIVDD126 pKa = 3.69QSTHH130 pKa = 6.39RR131 pKa = 11.84PVALLSGRR139 pKa = 11.84SATSLAEE146 pKa = 3.85WLKK149 pKa = 9.48TRR151 pKa = 11.84PEE153 pKa = 3.71IKK155 pKa = 9.96IITRR159 pKa = 11.84DD160 pKa = 3.25RR161 pKa = 11.84AGAYY165 pKa = 10.33ADD167 pKa = 3.64VAKK170 pKa = 9.96TGASQAIQIADD181 pKa = 3.6RR182 pKa = 11.84WHH184 pKa = 7.24LIRR187 pKa = 11.84NAGDD191 pKa = 3.02HH192 pKa = 6.38FEE194 pKa = 4.73RR195 pKa = 11.84FLKK198 pKa = 10.18RR199 pKa = 11.84HH200 pKa = 4.26WVAVKK205 pKa = 8.59EE206 pKa = 4.41TYY208 pKa = 9.89HH209 pKa = 7.08AIKK212 pKa = 8.68TQEE215 pKa = 3.78LSALQSEE222 pKa = 5.06PIITVSPQQEE232 pKa = 4.25TVTQQQRR239 pKa = 11.84RR240 pKa = 11.84LLFEE244 pKa = 5.2HH245 pKa = 7.2ISVLKK250 pKa = 9.93NKK252 pKa = 9.55GMAQTEE258 pKa = 4.08IALTAGISDD267 pKa = 3.43KK268 pKa = 9.25TVRR271 pKa = 11.84RR272 pKa = 11.84WLQTGGPPLCGKK284 pKa = 10.1RR285 pKa = 11.84PRR287 pKa = 11.84PRR289 pKa = 11.84QVIGFNEE296 pKa = 4.24QTWLRR301 pKa = 11.84EE302 pKa = 3.97QGNSGCHH309 pKa = 4.48NAALLFRR316 pKa = 11.84KK317 pKa = 9.82LKK319 pKa = 8.31TRR321 pKa = 11.84GYY323 pKa = 9.04VGGSATVRR331 pKa = 11.84SLVSQWRR338 pKa = 11.84YY339 pKa = 7.37TSAEE343 pKa = 4.07VLQYY347 pKa = 11.14PPMPRR352 pKa = 11.84LKK354 pKa = 10.36QVMQWLTSPLNLDD367 pKa = 3.36KK368 pKa = 11.34SGAATWIKK376 pKa = 10.71RR377 pKa = 11.84FVQQLLTLCPGLNVGCKK394 pKa = 8.46LTQGFISLFSPSTKK408 pKa = 9.13IKK410 pKa = 10.03EE411 pKa = 3.99RR412 pKa = 11.84SRR414 pKa = 11.84LLRR417 pKa = 11.84QWLHH421 pKa = 6.04EE422 pKa = 4.05ATHH425 pKa = 6.59CKK427 pKa = 10.12LSEE430 pKa = 4.06YY431 pKa = 10.86QKK433 pKa = 9.71MASSLEE439 pKa = 4.03MEE441 pKa = 4.88LDD443 pKa = 3.44AVEE446 pKa = 4.25NALTHH451 pKa = 6.01RR452 pKa = 11.84WSNGVVEE459 pKa = 4.11GHH461 pKa = 5.2VNRR464 pKa = 11.84LKK466 pKa = 9.78MLKK469 pKa = 9.26RR470 pKa = 11.84QMYY473 pKa = 8.84GRR475 pKa = 11.84AGFEE479 pKa = 3.89LLSRR483 pKa = 11.84RR484 pKa = 11.84VMNTLCC490 pKa = 4.7
MM1 pKa = 7.88LPQCNMKK8 pKa = 10.14SRR10 pKa = 11.84CRR12 pKa = 11.84YY13 pKa = 8.73GFTHH17 pKa = 7.19RR18 pKa = 11.84RR19 pKa = 11.84LQHH22 pKa = 6.15LPCGGKK28 pKa = 7.6TLILCFRR35 pKa = 11.84LQRR38 pKa = 11.84FCCTNSRR45 pKa = 11.84CPQRR49 pKa = 11.84TFQEE53 pKa = 4.68SISTLAPRR61 pKa = 11.84WQRR64 pKa = 11.84SSRR67 pKa = 11.84QLVEE71 pKa = 3.6MTTRR75 pKa = 11.84LAYY78 pKa = 10.42SFGVRR83 pKa = 11.84VSGDD87 pKa = 3.24TLIRR91 pKa = 11.84RR92 pKa = 11.84IMRR95 pKa = 11.84LPLSEE100 pKa = 4.78PEE102 pKa = 4.07QLTHH106 pKa = 7.61IGVDD110 pKa = 3.06DD111 pKa = 3.85WAWRR115 pKa = 11.84KK116 pKa = 8.0GHH118 pKa = 6.8HH119 pKa = 6.54YY120 pKa = 9.44GTIIVDD126 pKa = 3.69QSTHH130 pKa = 6.39RR131 pKa = 11.84PVALLSGRR139 pKa = 11.84SATSLAEE146 pKa = 3.85WLKK149 pKa = 9.48TRR151 pKa = 11.84PEE153 pKa = 3.71IKK155 pKa = 9.96IITRR159 pKa = 11.84DD160 pKa = 3.25RR161 pKa = 11.84AGAYY165 pKa = 10.33ADD167 pKa = 3.64VAKK170 pKa = 9.96TGASQAIQIADD181 pKa = 3.6RR182 pKa = 11.84WHH184 pKa = 7.24LIRR187 pKa = 11.84NAGDD191 pKa = 3.02HH192 pKa = 6.38FEE194 pKa = 4.73RR195 pKa = 11.84FLKK198 pKa = 10.18RR199 pKa = 11.84HH200 pKa = 4.26WVAVKK205 pKa = 8.59EE206 pKa = 4.41TYY208 pKa = 9.89HH209 pKa = 7.08AIKK212 pKa = 8.68TQEE215 pKa = 3.78LSALQSEE222 pKa = 5.06PIITVSPQQEE232 pKa = 4.25TVTQQQRR239 pKa = 11.84RR240 pKa = 11.84LLFEE244 pKa = 5.2HH245 pKa = 7.2ISVLKK250 pKa = 9.93NKK252 pKa = 9.55GMAQTEE258 pKa = 4.08IALTAGISDD267 pKa = 3.43KK268 pKa = 9.25TVRR271 pKa = 11.84RR272 pKa = 11.84WLQTGGPPLCGKK284 pKa = 10.1RR285 pKa = 11.84PRR287 pKa = 11.84PRR289 pKa = 11.84QVIGFNEE296 pKa = 4.24QTWLRR301 pKa = 11.84EE302 pKa = 3.97QGNSGCHH309 pKa = 4.48NAALLFRR316 pKa = 11.84KK317 pKa = 9.82LKK319 pKa = 8.31TRR321 pKa = 11.84GYY323 pKa = 9.04VGGSATVRR331 pKa = 11.84SLVSQWRR338 pKa = 11.84YY339 pKa = 7.37TSAEE343 pKa = 4.07VLQYY347 pKa = 11.14PPMPRR352 pKa = 11.84LKK354 pKa = 10.36QVMQWLTSPLNLDD367 pKa = 3.36KK368 pKa = 11.34SGAATWIKK376 pKa = 10.71RR377 pKa = 11.84FVQQLLTLCPGLNVGCKK394 pKa = 8.46LTQGFISLFSPSTKK408 pKa = 9.13IKK410 pKa = 10.03EE411 pKa = 3.99RR412 pKa = 11.84SRR414 pKa = 11.84LLRR417 pKa = 11.84QWLHH421 pKa = 6.04EE422 pKa = 4.05ATHH425 pKa = 6.59CKK427 pKa = 10.12LSEE430 pKa = 4.06YY431 pKa = 10.86QKK433 pKa = 9.71MASSLEE439 pKa = 4.03MEE441 pKa = 4.88LDD443 pKa = 3.44AVEE446 pKa = 4.25NALTHH451 pKa = 6.01RR452 pKa = 11.84WSNGVVEE459 pKa = 4.11GHH461 pKa = 5.2VNRR464 pKa = 11.84LKK466 pKa = 9.78MLKK469 pKa = 9.26RR470 pKa = 11.84QMYY473 pKa = 8.84GRR475 pKa = 11.84AGFEE479 pKa = 3.89LLSRR483 pKa = 11.84RR484 pKa = 11.84VMNTLCC490 pKa = 4.7
Molecular weight: 56.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
389023 |
30 |
1486 |
284.4 |
31.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.256 ± 0.058 | 1.211 ± 0.024 |
5.062 ± 0.055 | 5.758 ± 0.059 |
3.839 ± 0.044 | 6.241 ± 0.062 |
2.417 ± 0.033 | 7.11 ± 0.067 |
6.045 ± 0.059 | 10.789 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.601 ± 0.032 | 4.592 ± 0.046 |
4.058 ± 0.046 | 4.786 ± 0.056 |
5.338 ± 0.053 | 6.16 ± 0.051 |
5.347 ± 0.042 | 6.155 ± 0.055 |
1.21 ± 0.029 | 2.95 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
