
Rhynchobatus djiddensis polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
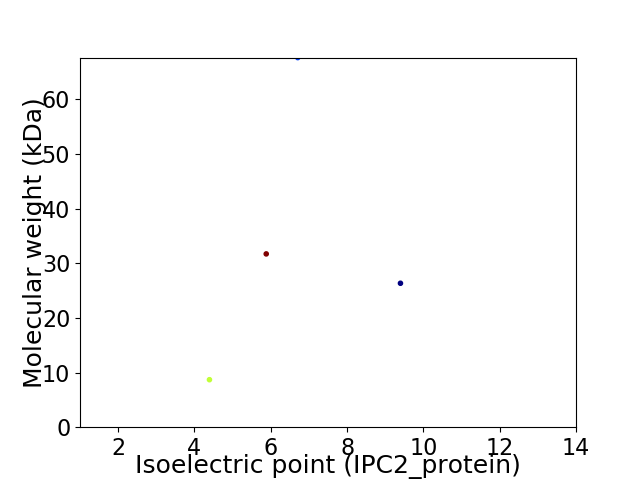
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5CVG5|A0A0B5CVG5_9POLY Large T OS=Rhynchobatus djiddensis polyomavirus 1 OX=2170102 PE=4 SV=1
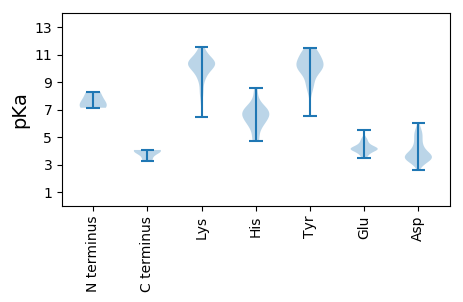
MM1 pKa = 7.67KK2 pKa = 10.42ARR4 pKa = 11.84GRR6 pKa = 11.84AFRR9 pKa = 11.84RR10 pKa = 11.84QLGLPCLCLNSKK22 pKa = 10.54PNVDD26 pKa = 4.36CDD28 pKa = 3.84VHH30 pKa = 8.06FCDD33 pKa = 6.01IDD35 pKa = 5.2DD36 pKa = 5.04DD37 pKa = 5.09SDD39 pKa = 3.44WDD41 pKa = 3.81SDD43 pKa = 3.37EE44 pKa = 5.48DD45 pKa = 4.73PIVNQVRR52 pKa = 11.84KK53 pKa = 10.34NIFDD57 pKa = 3.66NYY59 pKa = 8.78WRR61 pKa = 11.84DD62 pKa = 3.17RR63 pKa = 11.84LMCPEE68 pKa = 4.68VISDD72 pKa = 3.92SEE74 pKa = 4.06
MM1 pKa = 7.67KK2 pKa = 10.42ARR4 pKa = 11.84GRR6 pKa = 11.84AFRR9 pKa = 11.84RR10 pKa = 11.84QLGLPCLCLNSKK22 pKa = 10.54PNVDD26 pKa = 4.36CDD28 pKa = 3.84VHH30 pKa = 8.06FCDD33 pKa = 6.01IDD35 pKa = 5.2DD36 pKa = 5.04DD37 pKa = 5.09SDD39 pKa = 3.44WDD41 pKa = 3.81SDD43 pKa = 3.37EE44 pKa = 5.48DD45 pKa = 4.73PIVNQVRR52 pKa = 11.84KK53 pKa = 10.34NIFDD57 pKa = 3.66NYY59 pKa = 8.78WRR61 pKa = 11.84DD62 pKa = 3.17RR63 pKa = 11.84LMCPEE68 pKa = 4.68VISDD72 pKa = 3.92SEE74 pKa = 4.06
Molecular weight: 8.69 kDa
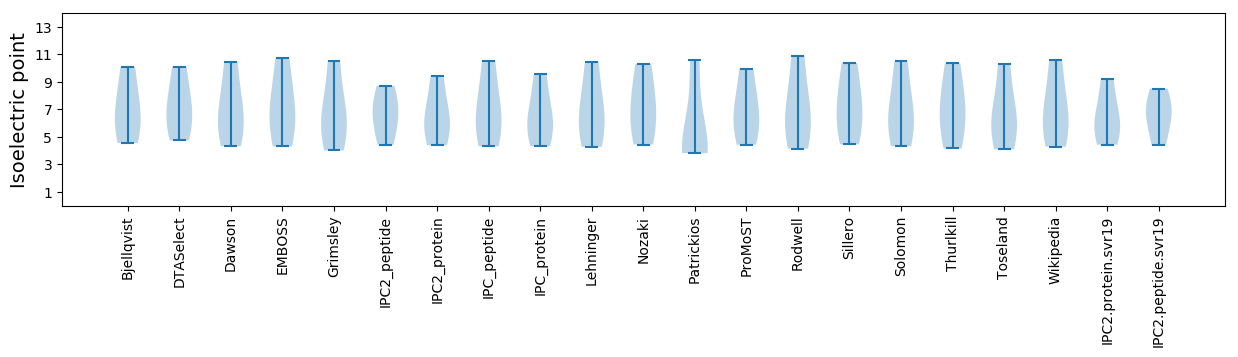
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5CSL6|A0A0B5CSL6_9POLY VP1 OS=Rhynchobatus djiddensis polyomavirus 1 OX=2170102 GN=VP1 PE=3 SV=1
MM1 pKa = 7.13GAVIAMLAAEE11 pKa = 5.04GGTLALEE18 pKa = 4.28EE19 pKa = 4.12ALGVSMAALEE29 pKa = 4.07TSAAIEE35 pKa = 4.16GFAGVGEE42 pKa = 4.17MMAASGMAGALEE54 pKa = 4.0EE55 pKa = 4.51AAMEE59 pKa = 4.25TLGTQAAQGMLSEE72 pKa = 4.5SLVGGGGIALATLNLSSGITGIMLSDD98 pKa = 3.26FHH100 pKa = 8.57GNGNRR105 pKa = 11.84FPYY108 pKa = 10.17LLKK111 pKa = 10.47EE112 pKa = 4.17NQEE115 pKa = 4.19GEE117 pKa = 4.15IPAVLLSLTKK127 pKa = 10.71SPIPGKK133 pKa = 9.96KK134 pKa = 7.55WGARR138 pKa = 11.84RR139 pKa = 11.84FVKK142 pKa = 10.43KK143 pKa = 10.35VGYY146 pKa = 8.84KK147 pKa = 9.69RR148 pKa = 11.84KK149 pKa = 9.81RR150 pKa = 11.84RR151 pKa = 11.84VKK153 pKa = 10.36KK154 pKa = 10.03RR155 pKa = 11.84AQPPQEE161 pKa = 3.95AEE163 pKa = 3.89EE164 pKa = 4.24VVEE167 pKa = 4.19EE168 pKa = 4.09EE169 pKa = 3.45RR170 pKa = 11.84RR171 pKa = 11.84YY172 pKa = 9.56PLRR175 pKa = 11.84RR176 pKa = 11.84GPLFRR181 pKa = 11.84RR182 pKa = 11.84PRR184 pKa = 11.84EE185 pKa = 3.73RR186 pKa = 11.84QPYY189 pKa = 9.28RR190 pKa = 11.84EE191 pKa = 4.28EE192 pKa = 3.9IEE194 pKa = 4.19NLLFVSAATSGVRR207 pKa = 11.84KK208 pKa = 9.34RR209 pKa = 11.84RR210 pKa = 11.84KK211 pKa = 9.05ARR213 pKa = 11.84KK214 pKa = 8.19PRR216 pKa = 11.84CTRR219 pKa = 11.84YY220 pKa = 9.93YY221 pKa = 10.38PNGNCQEE228 pKa = 4.24KK229 pKa = 10.26KK230 pKa = 10.5GPKK233 pKa = 9.5GPHH236 pKa = 4.8SSKK239 pKa = 10.51RR240 pKa = 11.84RR241 pKa = 11.84LL242 pKa = 3.29
MM1 pKa = 7.13GAVIAMLAAEE11 pKa = 5.04GGTLALEE18 pKa = 4.28EE19 pKa = 4.12ALGVSMAALEE29 pKa = 4.07TSAAIEE35 pKa = 4.16GFAGVGEE42 pKa = 4.17MMAASGMAGALEE54 pKa = 4.0EE55 pKa = 4.51AAMEE59 pKa = 4.25TLGTQAAQGMLSEE72 pKa = 4.5SLVGGGGIALATLNLSSGITGIMLSDD98 pKa = 3.26FHH100 pKa = 8.57GNGNRR105 pKa = 11.84FPYY108 pKa = 10.17LLKK111 pKa = 10.47EE112 pKa = 4.17NQEE115 pKa = 4.19GEE117 pKa = 4.15IPAVLLSLTKK127 pKa = 10.71SPIPGKK133 pKa = 9.96KK134 pKa = 7.55WGARR138 pKa = 11.84RR139 pKa = 11.84FVKK142 pKa = 10.43KK143 pKa = 10.35VGYY146 pKa = 8.84KK147 pKa = 9.69RR148 pKa = 11.84KK149 pKa = 9.81RR150 pKa = 11.84RR151 pKa = 11.84VKK153 pKa = 10.36KK154 pKa = 10.03RR155 pKa = 11.84AQPPQEE161 pKa = 3.95AEE163 pKa = 3.89EE164 pKa = 4.24VVEE167 pKa = 4.19EE168 pKa = 4.09EE169 pKa = 3.45RR170 pKa = 11.84RR171 pKa = 11.84YY172 pKa = 9.56PLRR175 pKa = 11.84RR176 pKa = 11.84GPLFRR181 pKa = 11.84RR182 pKa = 11.84PRR184 pKa = 11.84EE185 pKa = 3.73RR186 pKa = 11.84QPYY189 pKa = 9.28RR190 pKa = 11.84EE191 pKa = 4.28EE192 pKa = 3.9IEE194 pKa = 4.19NLLFVSAATSGVRR207 pKa = 11.84KK208 pKa = 9.34RR209 pKa = 11.84RR210 pKa = 11.84KK211 pKa = 9.05ARR213 pKa = 11.84KK214 pKa = 8.19PRR216 pKa = 11.84CTRR219 pKa = 11.84YY220 pKa = 9.93YY221 pKa = 10.38PNGNCQEE228 pKa = 4.24KK229 pKa = 10.26KK230 pKa = 10.5GPKK233 pKa = 9.5GPHH236 pKa = 4.8SSKK239 pKa = 10.51RR240 pKa = 11.84RR241 pKa = 11.84LL242 pKa = 3.29
Molecular weight: 26.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1190 |
74 |
597 |
297.5 |
33.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.387 ± 1.607 | 2.941 ± 0.939 |
5.798 ± 1.625 | 7.647 ± 0.869 |
4.286 ± 0.595 | 7.227 ± 1.359 |
2.101 ± 0.711 | 5.126 ± 0.612 |
7.479 ± 0.487 | 8.655 ± 0.463 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.941 ± 0.395 | 4.454 ± 0.443 |
4.79 ± 0.371 | 2.941 ± 0.052 |
6.723 ± 1.172 | 6.723 ± 0.435 |
4.538 ± 0.73 | 5.21 ± 0.295 |
0.84 ± 0.19 | 3.193 ± 0.37 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
