
Changjiang narna-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.0
Get precalculated fractions of proteins
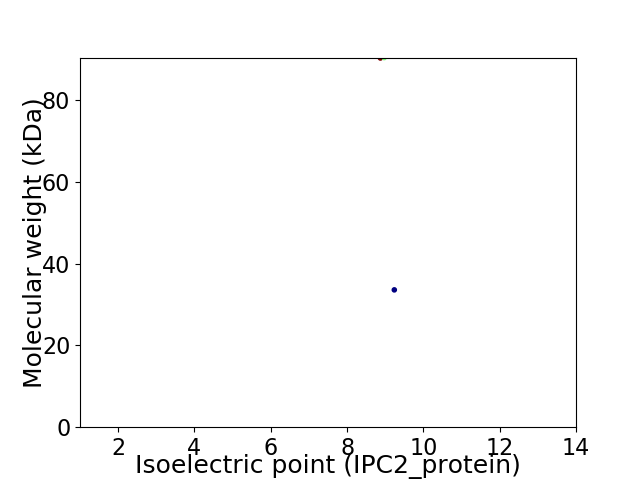
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
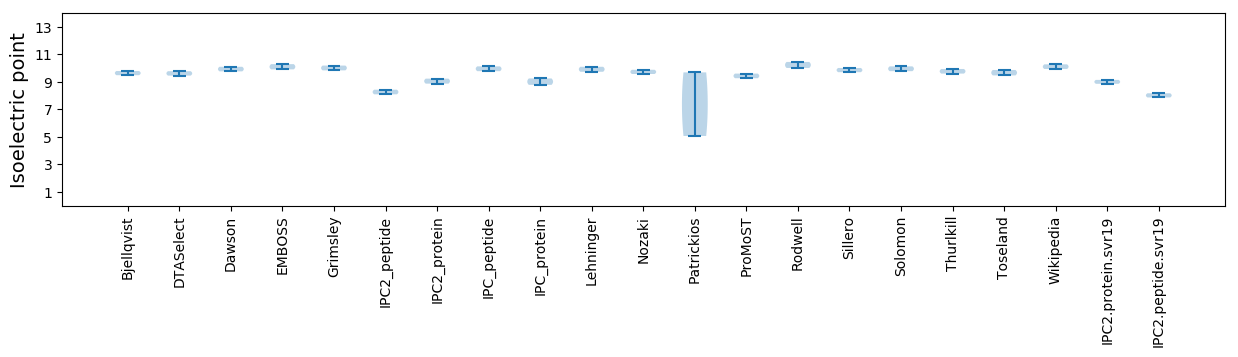
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KID5|A0A1L3KID5_9VIRU Uncharacterized protein OS=Changjiang narna-like virus 3 OX=1922778 PE=4 SV=1
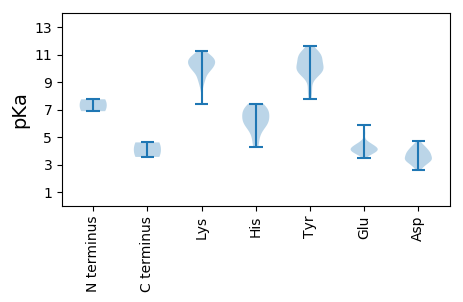
MM1 pKa = 6.88TRR3 pKa = 11.84NTITHH8 pKa = 5.86HH9 pKa = 5.42TAGGRR14 pKa = 11.84APQLSKK20 pKa = 10.9DD21 pKa = 3.35RR22 pKa = 11.84RR23 pKa = 11.84PRR25 pKa = 11.84RR26 pKa = 11.84VLRR29 pKa = 11.84STPTSADD36 pKa = 3.25LHH38 pKa = 6.12VLLPEE43 pKa = 4.36LVWDD47 pKa = 4.42PVQVARR53 pKa = 11.84LSAKK57 pKa = 10.03GCWRR61 pKa = 11.84KK62 pKa = 10.45ANDD65 pKa = 3.61ALIDD69 pKa = 3.75SFQLLGYY76 pKa = 9.69KK77 pKa = 9.84FEE79 pKa = 5.86NILCPVPPSSSVPEE93 pKa = 3.92LMSILKK99 pKa = 10.01SWTAFWLPHH108 pKa = 5.77MLGDD112 pKa = 3.86DD113 pKa = 3.84TPVQRR118 pKa = 11.84FNPYY122 pKa = 10.82SLITSEE128 pKa = 3.7FRR130 pKa = 11.84KK131 pKa = 10.19FIRR134 pKa = 11.84NRR136 pKa = 11.84VTGGGKK142 pKa = 9.11ARR144 pKa = 11.84KK145 pKa = 9.05YY146 pKa = 10.71RR147 pKa = 11.84IGALLLYY154 pKa = 9.99SKK156 pKa = 10.74RR157 pKa = 11.84LFPSFTVEE165 pKa = 3.84MVRR168 pKa = 11.84EE169 pKa = 4.02KK170 pKa = 11.21VNEE173 pKa = 3.88FSKK176 pKa = 10.79SVARR180 pKa = 11.84IEE182 pKa = 4.25PSVLPRR188 pKa = 11.84KK189 pKa = 9.62RR190 pKa = 11.84RR191 pKa = 11.84MFLEE195 pKa = 3.81IQRR198 pKa = 11.84TLDD201 pKa = 3.39EE202 pKa = 4.58FCPPGEE208 pKa = 4.21EE209 pKa = 4.36MVADD213 pKa = 3.79YY214 pKa = 9.85CKK216 pKa = 10.21PFPPSVSACHH226 pKa = 6.15EE227 pKa = 4.02YY228 pKa = 10.71SRR230 pKa = 11.84QEE232 pKa = 4.0GGLQAYY238 pKa = 9.26IRR240 pKa = 11.84DD241 pKa = 3.92FPLEE245 pKa = 4.27GFLEE249 pKa = 4.52DD250 pKa = 3.81PVVKK254 pKa = 10.56KK255 pKa = 10.48ILQQFRR261 pKa = 11.84LTDD264 pKa = 3.43TDD266 pKa = 3.98LSSNGPLNYY275 pKa = 9.84LWEE278 pKa = 4.45RR279 pKa = 11.84MLRR282 pKa = 11.84HH283 pKa = 6.72LIAEE287 pKa = 5.23AIGEE291 pKa = 4.19LGLPEE296 pKa = 5.26SEE298 pKa = 4.57WKK300 pKa = 10.4PMMVGATGLTEE311 pKa = 3.96PLKK314 pKa = 11.06VRR316 pKa = 11.84IVTKK320 pKa = 10.9AEE322 pKa = 3.72WFLQLLTPVQKK333 pKa = 10.46AWHH336 pKa = 5.77SKK338 pKa = 7.41MRR340 pKa = 11.84EE341 pKa = 3.92HH342 pKa = 7.43PVFQLIGGADD352 pKa = 3.43VEE354 pKa = 4.62DD355 pKa = 4.35ALRR358 pKa = 11.84PMKK361 pKa = 10.36LSKK364 pKa = 10.89GEE366 pKa = 4.04KK367 pKa = 9.46VVSGDD372 pKa = 3.39YY373 pKa = 10.73SAATDD378 pKa = 4.41NIFLTYY384 pKa = 9.56TKK386 pKa = 10.15EE387 pKa = 3.6AAEE390 pKa = 4.15AMLEE394 pKa = 4.01RR395 pKa = 11.84TRR397 pKa = 11.84FQLPEE402 pKa = 4.22SVPSYY407 pKa = 10.56TDD409 pKa = 2.78AFLRR413 pKa = 11.84KK414 pKa = 9.4LVVHH418 pKa = 6.67SLTKK422 pKa = 10.51SVLDD426 pKa = 4.23LKK428 pKa = 11.03GSSPVPITRR437 pKa = 11.84GQMMGHH443 pKa = 6.87ILSFPLLCIINRR455 pKa = 11.84AASCMAVSRR464 pKa = 11.84DD465 pKa = 2.83SFMRR469 pKa = 11.84INGDD473 pKa = 3.58DD474 pKa = 4.02VIFPATKK481 pKa = 9.91KK482 pKa = 10.92VYY484 pKa = 9.27TKK486 pKa = 9.62WKK488 pKa = 9.12SATKK492 pKa = 10.04IVGLEE497 pKa = 3.48FSIGKK502 pKa = 9.67NYY504 pKa = 10.09YY505 pKa = 10.34SRR507 pKa = 11.84DD508 pKa = 3.4LALVNSVYY516 pKa = 9.65CTYY519 pKa = 11.11SKK521 pKa = 11.26EE522 pKa = 3.82KK523 pKa = 10.67DD524 pKa = 2.6RR525 pKa = 11.84WIALDD530 pKa = 3.59VPNVGLLNMPLDD542 pKa = 3.8RR543 pKa = 11.84QVDD546 pKa = 4.33LNNGRR551 pKa = 11.84QILPWEE557 pKa = 3.93QLAQLFRR564 pKa = 11.84EE565 pKa = 4.73FNRR568 pKa = 11.84FSTSDD573 pKa = 2.91THH575 pKa = 6.28TKK577 pKa = 8.62YY578 pKa = 11.12LSMFRR583 pKa = 11.84KK584 pKa = 9.97YY585 pKa = 10.95YY586 pKa = 9.69PILRR590 pKa = 11.84GFPGPFYY597 pKa = 11.32GPVEE601 pKa = 4.18YY602 pKa = 10.35GAFGAPVPPKK612 pKa = 10.69HH613 pKa = 6.44EE614 pKa = 4.09FTKK617 pKa = 10.75NQLMWMNAHH626 pKa = 7.17RR627 pKa = 11.84LGIFNYY633 pKa = 10.11QEE635 pKa = 3.8GTRR638 pKa = 11.84NSFSKK643 pKa = 10.34ISNRR647 pKa = 11.84YY648 pKa = 7.79EE649 pKa = 3.75SFIAIEE655 pKa = 4.22LTKK658 pKa = 10.74GMYY661 pKa = 10.2KK662 pKa = 10.67FGPLPVGAAFGPPRR676 pKa = 11.84AADD679 pKa = 4.71GILDD683 pKa = 4.15PYY685 pKa = 11.12ARR687 pKa = 11.84DD688 pKa = 3.16GGLGYY693 pKa = 10.99RR694 pKa = 11.84LMAMRR699 pKa = 11.84RR700 pKa = 11.84WFEE703 pKa = 4.0DD704 pKa = 3.32LSSNKK709 pKa = 9.26HH710 pKa = 4.39VKK712 pKa = 10.1IFGARR717 pKa = 11.84RR718 pKa = 11.84WNHH721 pKa = 5.79FKK723 pKa = 11.17LSMKK727 pKa = 10.11DD728 pKa = 3.09QGGIPPLPPNFLHH741 pKa = 7.15KK742 pKa = 10.75VLEE745 pKa = 4.22NSTWSMRR752 pKa = 11.84PAWHH756 pKa = 6.71RR757 pKa = 11.84QRR759 pKa = 11.84DD760 pKa = 4.2TVGVRR765 pKa = 11.84YY766 pKa = 10.03KK767 pKa = 10.64EE768 pKa = 4.12DD769 pKa = 3.15ASFLHH774 pKa = 7.11EE775 pKa = 4.12IFQAQEE781 pKa = 3.79EE782 pKa = 4.49QTEE785 pKa = 4.44EE786 pKa = 3.98TTLL789 pKa = 3.55
MM1 pKa = 6.88TRR3 pKa = 11.84NTITHH8 pKa = 5.86HH9 pKa = 5.42TAGGRR14 pKa = 11.84APQLSKK20 pKa = 10.9DD21 pKa = 3.35RR22 pKa = 11.84RR23 pKa = 11.84PRR25 pKa = 11.84RR26 pKa = 11.84VLRR29 pKa = 11.84STPTSADD36 pKa = 3.25LHH38 pKa = 6.12VLLPEE43 pKa = 4.36LVWDD47 pKa = 4.42PVQVARR53 pKa = 11.84LSAKK57 pKa = 10.03GCWRR61 pKa = 11.84KK62 pKa = 10.45ANDD65 pKa = 3.61ALIDD69 pKa = 3.75SFQLLGYY76 pKa = 9.69KK77 pKa = 9.84FEE79 pKa = 5.86NILCPVPPSSSVPEE93 pKa = 3.92LMSILKK99 pKa = 10.01SWTAFWLPHH108 pKa = 5.77MLGDD112 pKa = 3.86DD113 pKa = 3.84TPVQRR118 pKa = 11.84FNPYY122 pKa = 10.82SLITSEE128 pKa = 3.7FRR130 pKa = 11.84KK131 pKa = 10.19FIRR134 pKa = 11.84NRR136 pKa = 11.84VTGGGKK142 pKa = 9.11ARR144 pKa = 11.84KK145 pKa = 9.05YY146 pKa = 10.71RR147 pKa = 11.84IGALLLYY154 pKa = 9.99SKK156 pKa = 10.74RR157 pKa = 11.84LFPSFTVEE165 pKa = 3.84MVRR168 pKa = 11.84EE169 pKa = 4.02KK170 pKa = 11.21VNEE173 pKa = 3.88FSKK176 pKa = 10.79SVARR180 pKa = 11.84IEE182 pKa = 4.25PSVLPRR188 pKa = 11.84KK189 pKa = 9.62RR190 pKa = 11.84RR191 pKa = 11.84MFLEE195 pKa = 3.81IQRR198 pKa = 11.84TLDD201 pKa = 3.39EE202 pKa = 4.58FCPPGEE208 pKa = 4.21EE209 pKa = 4.36MVADD213 pKa = 3.79YY214 pKa = 9.85CKK216 pKa = 10.21PFPPSVSACHH226 pKa = 6.15EE227 pKa = 4.02YY228 pKa = 10.71SRR230 pKa = 11.84QEE232 pKa = 4.0GGLQAYY238 pKa = 9.26IRR240 pKa = 11.84DD241 pKa = 3.92FPLEE245 pKa = 4.27GFLEE249 pKa = 4.52DD250 pKa = 3.81PVVKK254 pKa = 10.56KK255 pKa = 10.48ILQQFRR261 pKa = 11.84LTDD264 pKa = 3.43TDD266 pKa = 3.98LSSNGPLNYY275 pKa = 9.84LWEE278 pKa = 4.45RR279 pKa = 11.84MLRR282 pKa = 11.84HH283 pKa = 6.72LIAEE287 pKa = 5.23AIGEE291 pKa = 4.19LGLPEE296 pKa = 5.26SEE298 pKa = 4.57WKK300 pKa = 10.4PMMVGATGLTEE311 pKa = 3.96PLKK314 pKa = 11.06VRR316 pKa = 11.84IVTKK320 pKa = 10.9AEE322 pKa = 3.72WFLQLLTPVQKK333 pKa = 10.46AWHH336 pKa = 5.77SKK338 pKa = 7.41MRR340 pKa = 11.84EE341 pKa = 3.92HH342 pKa = 7.43PVFQLIGGADD352 pKa = 3.43VEE354 pKa = 4.62DD355 pKa = 4.35ALRR358 pKa = 11.84PMKK361 pKa = 10.36LSKK364 pKa = 10.89GEE366 pKa = 4.04KK367 pKa = 9.46VVSGDD372 pKa = 3.39YY373 pKa = 10.73SAATDD378 pKa = 4.41NIFLTYY384 pKa = 9.56TKK386 pKa = 10.15EE387 pKa = 3.6AAEE390 pKa = 4.15AMLEE394 pKa = 4.01RR395 pKa = 11.84TRR397 pKa = 11.84FQLPEE402 pKa = 4.22SVPSYY407 pKa = 10.56TDD409 pKa = 2.78AFLRR413 pKa = 11.84KK414 pKa = 9.4LVVHH418 pKa = 6.67SLTKK422 pKa = 10.51SVLDD426 pKa = 4.23LKK428 pKa = 11.03GSSPVPITRR437 pKa = 11.84GQMMGHH443 pKa = 6.87ILSFPLLCIINRR455 pKa = 11.84AASCMAVSRR464 pKa = 11.84DD465 pKa = 2.83SFMRR469 pKa = 11.84INGDD473 pKa = 3.58DD474 pKa = 4.02VIFPATKK481 pKa = 9.91KK482 pKa = 10.92VYY484 pKa = 9.27TKK486 pKa = 9.62WKK488 pKa = 9.12SATKK492 pKa = 10.04IVGLEE497 pKa = 3.48FSIGKK502 pKa = 9.67NYY504 pKa = 10.09YY505 pKa = 10.34SRR507 pKa = 11.84DD508 pKa = 3.4LALVNSVYY516 pKa = 9.65CTYY519 pKa = 11.11SKK521 pKa = 11.26EE522 pKa = 3.82KK523 pKa = 10.67DD524 pKa = 2.6RR525 pKa = 11.84WIALDD530 pKa = 3.59VPNVGLLNMPLDD542 pKa = 3.8RR543 pKa = 11.84QVDD546 pKa = 4.33LNNGRR551 pKa = 11.84QILPWEE557 pKa = 3.93QLAQLFRR564 pKa = 11.84EE565 pKa = 4.73FNRR568 pKa = 11.84FSTSDD573 pKa = 2.91THH575 pKa = 6.28TKK577 pKa = 8.62YY578 pKa = 11.12LSMFRR583 pKa = 11.84KK584 pKa = 9.97YY585 pKa = 10.95YY586 pKa = 9.69PILRR590 pKa = 11.84GFPGPFYY597 pKa = 11.32GPVEE601 pKa = 4.18YY602 pKa = 10.35GAFGAPVPPKK612 pKa = 10.69HH613 pKa = 6.44EE614 pKa = 4.09FTKK617 pKa = 10.75NQLMWMNAHH626 pKa = 7.17RR627 pKa = 11.84LGIFNYY633 pKa = 10.11QEE635 pKa = 3.8GTRR638 pKa = 11.84NSFSKK643 pKa = 10.34ISNRR647 pKa = 11.84YY648 pKa = 7.79EE649 pKa = 3.75SFIAIEE655 pKa = 4.22LTKK658 pKa = 10.74GMYY661 pKa = 10.2KK662 pKa = 10.67FGPLPVGAAFGPPRR676 pKa = 11.84AADD679 pKa = 4.71GILDD683 pKa = 4.15PYY685 pKa = 11.12ARR687 pKa = 11.84DD688 pKa = 3.16GGLGYY693 pKa = 10.99RR694 pKa = 11.84LMAMRR699 pKa = 11.84RR700 pKa = 11.84WFEE703 pKa = 4.0DD704 pKa = 3.32LSSNKK709 pKa = 9.26HH710 pKa = 4.39VKK712 pKa = 10.1IFGARR717 pKa = 11.84RR718 pKa = 11.84WNHH721 pKa = 5.79FKK723 pKa = 11.17LSMKK727 pKa = 10.11DD728 pKa = 3.09QGGIPPLPPNFLHH741 pKa = 7.15KK742 pKa = 10.75VLEE745 pKa = 4.22NSTWSMRR752 pKa = 11.84PAWHH756 pKa = 6.71RR757 pKa = 11.84QRR759 pKa = 11.84DD760 pKa = 4.2TVGVRR765 pKa = 11.84YY766 pKa = 10.03KK767 pKa = 10.64EE768 pKa = 4.12DD769 pKa = 3.15ASFLHH774 pKa = 7.11EE775 pKa = 4.12IFQAQEE781 pKa = 3.79EE782 pKa = 4.49QTEE785 pKa = 4.44EE786 pKa = 3.98TTLL789 pKa = 3.55
Molecular weight: 90.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KID5|A0A1L3KID5_9VIRU Uncharacterized protein OS=Changjiang narna-like virus 3 OX=1922778 PE=4 SV=1
MM1 pKa = 7.75AKK3 pKa = 9.78TNTKK7 pKa = 10.36KK8 pKa = 10.53SGKK11 pKa = 10.17KK12 pKa = 8.99NVKK15 pKa = 9.89NSNTGNKK22 pKa = 8.5PRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TMGGGSVAGRR36 pKa = 11.84YY37 pKa = 8.31AQLLHH42 pKa = 6.66NPDD45 pKa = 4.05NGDD48 pKa = 3.26HH49 pKa = 6.28MFDD52 pKa = 3.39VYY54 pKa = 11.02DD55 pKa = 3.82GEE57 pKa = 4.49RR58 pKa = 11.84GEE60 pKa = 4.35TQKK63 pKa = 10.63FVSTITLNTIAGHH76 pKa = 4.98TAGFLQLFGATGNGQWQSGASSSAALTFALLNTGCPGAAYY116 pKa = 8.48LTANAAKK123 pKa = 10.08SRR125 pKa = 11.84CKK127 pKa = 10.31ALKK130 pKa = 10.67LEE132 pKa = 5.59LIPAAASFSNITGEE146 pKa = 4.09VAAGVTTSGSWASGVTSVDD165 pKa = 3.21QLFDD169 pKa = 2.98IAKK172 pKa = 10.04AYY174 pKa = 10.34GPLRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.22TVVSRR185 pKa = 11.84WIPSGLDD192 pKa = 3.02HH193 pKa = 7.33TYY195 pKa = 11.59ANYY198 pKa = 10.87NSVPNEE204 pKa = 4.04DD205 pKa = 3.64SNAVYY210 pKa = 9.89IAYY213 pKa = 9.86RR214 pKa = 11.84GWPAGVPISIRR225 pKa = 11.84MTYY228 pKa = 9.78VVEE231 pKa = 4.19FTIKK235 pKa = 10.49NSIGIPPTGAVSVPVGHH252 pKa = 5.96PHH254 pKa = 6.92ILAAMQQQDD263 pKa = 4.06PHH265 pKa = 5.09WHH267 pKa = 6.22HH268 pKa = 7.13SILDD272 pKa = 3.64EE273 pKa = 4.3VKK275 pKa = 9.74RR276 pKa = 11.84AGVGVARR283 pKa = 11.84DD284 pKa = 3.39VGNFTRR290 pKa = 11.84HH291 pKa = 4.31MARR294 pKa = 11.84NGMVRR299 pKa = 11.84VAEE302 pKa = 4.4KK303 pKa = 10.31VFSRR307 pKa = 11.84GVSSAIVLAA316 pKa = 4.61
MM1 pKa = 7.75AKK3 pKa = 9.78TNTKK7 pKa = 10.36KK8 pKa = 10.53SGKK11 pKa = 10.17KK12 pKa = 8.99NVKK15 pKa = 9.89NSNTGNKK22 pKa = 8.5PRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TMGGGSVAGRR36 pKa = 11.84YY37 pKa = 8.31AQLLHH42 pKa = 6.66NPDD45 pKa = 4.05NGDD48 pKa = 3.26HH49 pKa = 6.28MFDD52 pKa = 3.39VYY54 pKa = 11.02DD55 pKa = 3.82GEE57 pKa = 4.49RR58 pKa = 11.84GEE60 pKa = 4.35TQKK63 pKa = 10.63FVSTITLNTIAGHH76 pKa = 4.98TAGFLQLFGATGNGQWQSGASSSAALTFALLNTGCPGAAYY116 pKa = 8.48LTANAAKK123 pKa = 10.08SRR125 pKa = 11.84CKK127 pKa = 10.31ALKK130 pKa = 10.67LEE132 pKa = 5.59LIPAAASFSNITGEE146 pKa = 4.09VAAGVTTSGSWASGVTSVDD165 pKa = 3.21QLFDD169 pKa = 2.98IAKK172 pKa = 10.04AYY174 pKa = 10.34GPLRR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 4.22TVVSRR185 pKa = 11.84WIPSGLDD192 pKa = 3.02HH193 pKa = 7.33TYY195 pKa = 11.59ANYY198 pKa = 10.87NSVPNEE204 pKa = 4.04DD205 pKa = 3.64SNAVYY210 pKa = 9.89IAYY213 pKa = 9.86RR214 pKa = 11.84GWPAGVPISIRR225 pKa = 11.84MTYY228 pKa = 9.78VVEE231 pKa = 4.19FTIKK235 pKa = 10.49NSIGIPPTGAVSVPVGHH252 pKa = 5.96PHH254 pKa = 6.92ILAAMQQQDD263 pKa = 4.06PHH265 pKa = 5.09WHH267 pKa = 6.22HH268 pKa = 7.13SILDD272 pKa = 3.64EE273 pKa = 4.3VKK275 pKa = 9.74RR276 pKa = 11.84AGVGVARR283 pKa = 11.84DD284 pKa = 3.39VGNFTRR290 pKa = 11.84HH291 pKa = 4.31MARR294 pKa = 11.84NGMVRR299 pKa = 11.84VAEE302 pKa = 4.4KK303 pKa = 10.31VFSRR307 pKa = 11.84GVSSAIVLAA316 pKa = 4.61
Molecular weight: 33.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1105 |
316 |
789 |
552.5 |
61.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.783 ± 2.025 | 0.905 ± 0.14 |
4.163 ± 0.352 | 5.068 ± 1.145 |
4.796 ± 0.841 | 7.421 ± 1.558 |
2.534 ± 0.325 | 4.525 ± 0.114 |
5.701 ± 0.492 | 8.959 ± 1.683 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.896 ± 0.351 | 4.163 ± 0.954 |
6.244 ± 0.935 | 3.077 ± 0.118 |
6.878 ± 0.773 | 7.33 ± 0.463 |
5.701 ± 0.813 | 6.697 ± 0.953 |
1.9 ± 0.164 | 3.258 ± 0.211 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
