
Haloarculaceae archaeon HArcel1
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; unclassified Haloarculaceae
Average proteome isoelectric point is 4.85
Get precalculated fractions of proteins
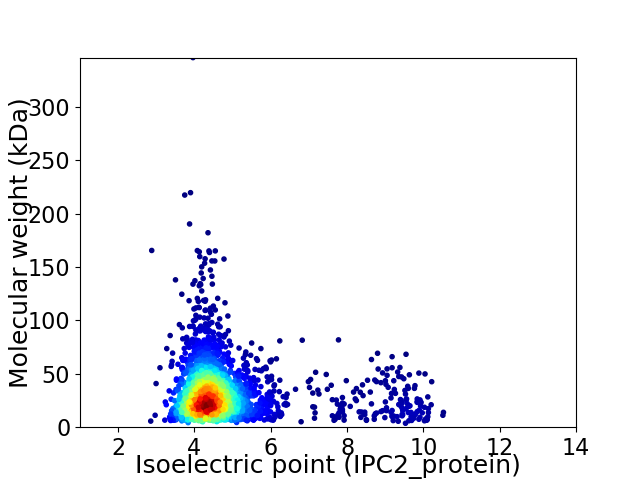
Virtual 2D-PAGE plot for 2532 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R4X102|A0A2R4X102_9EURY Uncharacterized protein OS=Haloarculaceae archaeon HArcel1 OX=1679096 GN=HARCEL1_06995 PE=4 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84SPEE5 pKa = 3.75VDD7 pKa = 3.13RR8 pKa = 11.84QQVARR13 pKa = 11.84MQQSPTRR20 pKa = 11.84RR21 pKa = 11.84QILRR25 pKa = 11.84ASGATILGGSVLTAGTHH42 pKa = 4.47TVTAHH47 pKa = 7.27RR48 pKa = 11.84STDD51 pKa = 3.0DD52 pKa = 2.94WHH54 pKa = 6.9QFGYY58 pKa = 11.22DD59 pKa = 3.58DD60 pKa = 5.93ANTGHH65 pKa = 7.35AADD68 pKa = 3.81TTGPVTGVSPEE79 pKa = 3.5WHH81 pKa = 6.55FPDD84 pKa = 4.0VSGLEE89 pKa = 4.13SSPTADD95 pKa = 3.3GDD97 pKa = 4.13TVYY100 pKa = 10.84LGTVRR105 pKa = 11.84GVVALSEE112 pKa = 4.33ADD114 pKa = 3.4RR115 pKa = 11.84SVRR118 pKa = 11.84WRR120 pKa = 11.84TRR122 pKa = 11.84LSSPVKK128 pKa = 7.52TTPVLANGTLYY139 pKa = 11.04VGTADD144 pKa = 3.66GTVCAVDD151 pKa = 3.67PASGAVQWRR160 pKa = 11.84FEE162 pKa = 4.34TNARR166 pKa = 11.84MYY168 pKa = 9.48FSPVVSDD175 pKa = 3.1GTVYY179 pKa = 10.71VGSGDD184 pKa = 3.52ASVYY188 pKa = 11.08ALDD191 pKa = 3.95AVEE194 pKa = 5.52GTEE197 pKa = 3.64LWQYY201 pKa = 7.02EE202 pKa = 4.35TGAEE206 pKa = 4.34VHH208 pKa = 6.01CTPAVADD215 pKa = 3.62GTVYY219 pKa = 10.94VGGIDD224 pKa = 3.66NNVYY228 pKa = 10.83ALDD231 pKa = 4.37AGDD234 pKa = 5.21GTVEE238 pKa = 4.02WQFEE242 pKa = 4.47TEE244 pKa = 4.17DD245 pKa = 3.72TVMGSPAIADD255 pKa = 3.43GTVYY259 pKa = 10.65VGSFDD264 pKa = 3.58NTVYY268 pKa = 10.91AVDD271 pKa = 3.43ASGGSEE277 pKa = 3.66EE278 pKa = 4.15WRR280 pKa = 11.84FDD282 pKa = 2.96TGDD285 pKa = 3.56RR286 pKa = 11.84VNSSPAVADD295 pKa = 3.38GTVFVGSFDD304 pKa = 4.11NNVYY308 pKa = 10.92ALDD311 pKa = 3.93ARR313 pKa = 11.84DD314 pKa = 4.08GAVEE318 pKa = 3.96WRR320 pKa = 11.84FEE322 pKa = 3.76TDD324 pKa = 2.5DD325 pKa = 3.85WVNSSPVIADD335 pKa = 3.35GTVHH339 pKa = 6.13VGSYY343 pKa = 10.52DD344 pKa = 3.58GNVYY348 pKa = 10.87ALDD351 pKa = 3.83RR352 pKa = 11.84THH354 pKa = 7.06GAVKK358 pKa = 9.6WRR360 pKa = 11.84FEE362 pKa = 3.95TGGWEE367 pKa = 3.98RR368 pKa = 11.84SQSAVADD375 pKa = 4.09GTISPSVRR383 pKa = 11.84DD384 pKa = 3.31STVYY388 pKa = 10.73VGTQNSVYY396 pKa = 10.62AIDD399 pKa = 4.01AANGTEE405 pKa = 3.53RR406 pKa = 11.84WQYY409 pKa = 8.39EE410 pKa = 3.93TGAPNDD416 pKa = 3.85SSPAAVDD423 pKa = 3.33GTVYY427 pKa = 10.78LGSTNSRR434 pKa = 11.84VSAIDD439 pKa = 3.62AGDD442 pKa = 3.81GTVQWQFEE450 pKa = 4.48TGDD453 pKa = 3.96EE454 pKa = 4.43VHH456 pKa = 6.85SSPAVDD462 pKa = 4.88DD463 pKa = 3.89GTVYY467 pKa = 10.37IGSWDD472 pKa = 3.54NRR474 pKa = 11.84LYY476 pKa = 11.25ALDD479 pKa = 4.15VDD481 pKa = 5.1DD482 pKa = 6.4GTIQWRR488 pKa = 11.84FEE490 pKa = 3.95TDD492 pKa = 3.13GEE494 pKa = 4.83LNCSPVVADD503 pKa = 3.29GTVYY507 pKa = 10.68VGSLDD512 pKa = 3.54NHH514 pKa = 6.61VYY516 pKa = 10.6AVDD519 pKa = 3.98ADD521 pKa = 4.24DD522 pKa = 3.9GTEE525 pKa = 3.42RR526 pKa = 11.84WRR528 pKa = 11.84YY529 pKa = 5.17EE530 pKa = 3.93TGDD533 pKa = 3.76EE534 pKa = 4.36VHH536 pKa = 6.85SSPAIVDD543 pKa = 3.38GTVYY547 pKa = 10.54VGSWDD552 pKa = 3.64DD553 pKa = 3.65SIYY556 pKa = 10.87AIDD559 pKa = 4.53AGDD562 pKa = 3.7GTVQWRR568 pKa = 11.84YY569 pKa = 6.17EE570 pKa = 4.14TTGGVSSSPAVANDD584 pKa = 3.46TVYY587 pKa = 10.94VGNFEE592 pKa = 4.69GIVYY596 pKa = 10.65ALDD599 pKa = 4.27AEE601 pKa = 4.79DD602 pKa = 6.07GSVEE606 pKa = 3.82WGQTTNGVINSSPAIADD623 pKa = 3.42TTVYY627 pKa = 10.05IGSWDD632 pKa = 3.59NTVYY636 pKa = 11.21ALDD639 pKa = 4.85ADD641 pKa = 4.75DD642 pKa = 4.14GTKK645 pKa = 9.74RR646 pKa = 11.84WEE648 pKa = 3.85YY649 pKa = 9.73RR650 pKa = 11.84TDD652 pKa = 3.97DD653 pKa = 4.91EE654 pKa = 4.63IHH656 pKa = 6.71SSPAVADD663 pKa = 3.49GTVYY667 pKa = 10.71VGSDD671 pKa = 3.64DD672 pKa = 4.28DD673 pKa = 4.65SVYY676 pKa = 11.37ALDD679 pKa = 4.06ATDD682 pKa = 5.49GTVQWQYY689 pKa = 9.74EE690 pKa = 4.23TGDD693 pKa = 3.72WVPQSPSVADD703 pKa = 3.4GTVYY707 pKa = 10.32IGSFDD712 pKa = 3.62GLVYY716 pKa = 11.0ALTEE720 pKa = 4.15PARR723 pKa = 11.84DD724 pKa = 3.71TPSPAVEE731 pKa = 4.09PTEE734 pKa = 4.09TAEE737 pKa = 4.46PRR739 pKa = 11.84QQDD742 pKa = 3.86TEE744 pKa = 4.33GSGIDD749 pKa = 4.12ALPIAAGLGLIGLGGGGGALWWVRR773 pKa = 11.84QQ774 pKa = 3.57
MM1 pKa = 8.02RR2 pKa = 11.84SPEE5 pKa = 3.75VDD7 pKa = 3.13RR8 pKa = 11.84QQVARR13 pKa = 11.84MQQSPTRR20 pKa = 11.84RR21 pKa = 11.84QILRR25 pKa = 11.84ASGATILGGSVLTAGTHH42 pKa = 4.47TVTAHH47 pKa = 7.27RR48 pKa = 11.84STDD51 pKa = 3.0DD52 pKa = 2.94WHH54 pKa = 6.9QFGYY58 pKa = 11.22DD59 pKa = 3.58DD60 pKa = 5.93ANTGHH65 pKa = 7.35AADD68 pKa = 3.81TTGPVTGVSPEE79 pKa = 3.5WHH81 pKa = 6.55FPDD84 pKa = 4.0VSGLEE89 pKa = 4.13SSPTADD95 pKa = 3.3GDD97 pKa = 4.13TVYY100 pKa = 10.84LGTVRR105 pKa = 11.84GVVALSEE112 pKa = 4.33ADD114 pKa = 3.4RR115 pKa = 11.84SVRR118 pKa = 11.84WRR120 pKa = 11.84TRR122 pKa = 11.84LSSPVKK128 pKa = 7.52TTPVLANGTLYY139 pKa = 11.04VGTADD144 pKa = 3.66GTVCAVDD151 pKa = 3.67PASGAVQWRR160 pKa = 11.84FEE162 pKa = 4.34TNARR166 pKa = 11.84MYY168 pKa = 9.48FSPVVSDD175 pKa = 3.1GTVYY179 pKa = 10.71VGSGDD184 pKa = 3.52ASVYY188 pKa = 11.08ALDD191 pKa = 3.95AVEE194 pKa = 5.52GTEE197 pKa = 3.64LWQYY201 pKa = 7.02EE202 pKa = 4.35TGAEE206 pKa = 4.34VHH208 pKa = 6.01CTPAVADD215 pKa = 3.62GTVYY219 pKa = 10.94VGGIDD224 pKa = 3.66NNVYY228 pKa = 10.83ALDD231 pKa = 4.37AGDD234 pKa = 5.21GTVEE238 pKa = 4.02WQFEE242 pKa = 4.47TEE244 pKa = 4.17DD245 pKa = 3.72TVMGSPAIADD255 pKa = 3.43GTVYY259 pKa = 10.65VGSFDD264 pKa = 3.58NTVYY268 pKa = 10.91AVDD271 pKa = 3.43ASGGSEE277 pKa = 3.66EE278 pKa = 4.15WRR280 pKa = 11.84FDD282 pKa = 2.96TGDD285 pKa = 3.56RR286 pKa = 11.84VNSSPAVADD295 pKa = 3.38GTVFVGSFDD304 pKa = 4.11NNVYY308 pKa = 10.92ALDD311 pKa = 3.93ARR313 pKa = 11.84DD314 pKa = 4.08GAVEE318 pKa = 3.96WRR320 pKa = 11.84FEE322 pKa = 3.76TDD324 pKa = 2.5DD325 pKa = 3.85WVNSSPVIADD335 pKa = 3.35GTVHH339 pKa = 6.13VGSYY343 pKa = 10.52DD344 pKa = 3.58GNVYY348 pKa = 10.87ALDD351 pKa = 3.83RR352 pKa = 11.84THH354 pKa = 7.06GAVKK358 pKa = 9.6WRR360 pKa = 11.84FEE362 pKa = 3.95TGGWEE367 pKa = 3.98RR368 pKa = 11.84SQSAVADD375 pKa = 4.09GTISPSVRR383 pKa = 11.84DD384 pKa = 3.31STVYY388 pKa = 10.73VGTQNSVYY396 pKa = 10.62AIDD399 pKa = 4.01AANGTEE405 pKa = 3.53RR406 pKa = 11.84WQYY409 pKa = 8.39EE410 pKa = 3.93TGAPNDD416 pKa = 3.85SSPAAVDD423 pKa = 3.33GTVYY427 pKa = 10.78LGSTNSRR434 pKa = 11.84VSAIDD439 pKa = 3.62AGDD442 pKa = 3.81GTVQWQFEE450 pKa = 4.48TGDD453 pKa = 3.96EE454 pKa = 4.43VHH456 pKa = 6.85SSPAVDD462 pKa = 4.88DD463 pKa = 3.89GTVYY467 pKa = 10.37IGSWDD472 pKa = 3.54NRR474 pKa = 11.84LYY476 pKa = 11.25ALDD479 pKa = 4.15VDD481 pKa = 5.1DD482 pKa = 6.4GTIQWRR488 pKa = 11.84FEE490 pKa = 3.95TDD492 pKa = 3.13GEE494 pKa = 4.83LNCSPVVADD503 pKa = 3.29GTVYY507 pKa = 10.68VGSLDD512 pKa = 3.54NHH514 pKa = 6.61VYY516 pKa = 10.6AVDD519 pKa = 3.98ADD521 pKa = 4.24DD522 pKa = 3.9GTEE525 pKa = 3.42RR526 pKa = 11.84WRR528 pKa = 11.84YY529 pKa = 5.17EE530 pKa = 3.93TGDD533 pKa = 3.76EE534 pKa = 4.36VHH536 pKa = 6.85SSPAIVDD543 pKa = 3.38GTVYY547 pKa = 10.54VGSWDD552 pKa = 3.64DD553 pKa = 3.65SIYY556 pKa = 10.87AIDD559 pKa = 4.53AGDD562 pKa = 3.7GTVQWRR568 pKa = 11.84YY569 pKa = 6.17EE570 pKa = 4.14TTGGVSSSPAVANDD584 pKa = 3.46TVYY587 pKa = 10.94VGNFEE592 pKa = 4.69GIVYY596 pKa = 10.65ALDD599 pKa = 4.27AEE601 pKa = 4.79DD602 pKa = 6.07GSVEE606 pKa = 3.82WGQTTNGVINSSPAIADD623 pKa = 3.42TTVYY627 pKa = 10.05IGSWDD632 pKa = 3.59NTVYY636 pKa = 11.21ALDD639 pKa = 4.85ADD641 pKa = 4.75DD642 pKa = 4.14GTKK645 pKa = 9.74RR646 pKa = 11.84WEE648 pKa = 3.85YY649 pKa = 9.73RR650 pKa = 11.84TDD652 pKa = 3.97DD653 pKa = 4.91EE654 pKa = 4.63IHH656 pKa = 6.71SSPAVADD663 pKa = 3.49GTVYY667 pKa = 10.71VGSDD671 pKa = 3.64DD672 pKa = 4.28DD673 pKa = 4.65SVYY676 pKa = 11.37ALDD679 pKa = 4.06ATDD682 pKa = 5.49GTVQWQYY689 pKa = 9.74EE690 pKa = 4.23TGDD693 pKa = 3.72WVPQSPSVADD703 pKa = 3.4GTVYY707 pKa = 10.32IGSFDD712 pKa = 3.62GLVYY716 pKa = 11.0ALTEE720 pKa = 4.15PARR723 pKa = 11.84DD724 pKa = 3.71TPSPAVEE731 pKa = 4.09PTEE734 pKa = 4.09TAEE737 pKa = 4.46PRR739 pKa = 11.84QQDD742 pKa = 3.86TEE744 pKa = 4.33GSGIDD749 pKa = 4.12ALPIAAGLGLIGLGGGGGALWWVRR773 pKa = 11.84QQ774 pKa = 3.57
Molecular weight: 82.25 kDa
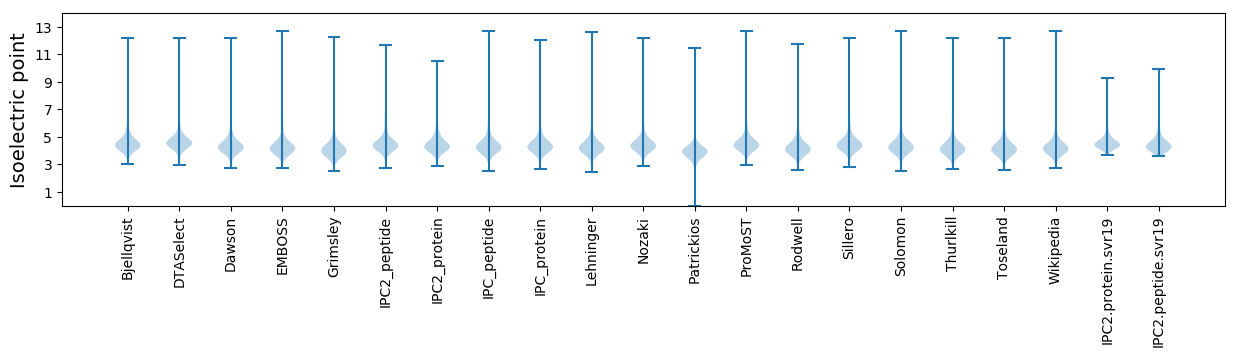
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R4X1I0|A0A2R4X1I0_9EURY Uncharacterized protein OS=Haloarculaceae archaeon HArcel1 OX=1679096 GN=HARCEL1_07955 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84ISTAVSWLVVCAGVAALGAVVWTDD26 pKa = 3.16RR27 pKa = 11.84VARR30 pKa = 11.84AVVPRR35 pKa = 11.84AVGAGLLALLVGVTLGRR52 pKa = 11.84LASTRR57 pKa = 11.84TRR59 pKa = 11.84QASTGLGAPRR69 pKa = 11.84VWPVIVIGGVLGSAVLATIGALGGGLVALPIVEE102 pKa = 4.72APLTVEE108 pKa = 4.57ALRR111 pKa = 11.84TTPLVRR117 pKa = 11.84RR118 pKa = 11.84AALAGVVLTHH128 pKa = 6.96ALAGAAGLTTLAHH141 pKa = 7.0TITPGDD147 pKa = 3.35RR148 pKa = 11.84VAATGPAISLAAIAAAGLALAPIGTVPMAGLTVALGALGVGLYY191 pKa = 10.48ARR193 pKa = 11.84APIRR197 pKa = 11.84ALLDD201 pKa = 3.46RR202 pKa = 11.84LGLDD206 pKa = 3.4ARR208 pKa = 11.84VLPARR213 pKa = 11.84SEE215 pKa = 4.16WAHH218 pKa = 6.55DD219 pKa = 3.5AADD222 pKa = 3.45AEE224 pKa = 4.54RR225 pKa = 11.84AHH227 pKa = 7.2RR228 pKa = 11.84RR229 pKa = 11.84AQKK232 pKa = 10.07RR233 pKa = 11.84APPLDD238 pKa = 3.43IGEE241 pKa = 4.37TTSIVVQEE249 pKa = 4.22RR250 pKa = 11.84LDD252 pKa = 3.48GGRR255 pKa = 11.84QLRR258 pKa = 11.84GVYY261 pKa = 10.26EE262 pKa = 4.2DD263 pKa = 3.62FQVFVKK269 pKa = 10.18WDD271 pKa = 3.75VPPDD275 pKa = 3.66CATGEE280 pKa = 4.45TICVRR285 pKa = 11.84IEE287 pKa = 4.23DD288 pKa = 4.44YY289 pKa = 9.47GTNDD293 pKa = 2.76AGQRR297 pKa = 11.84TSGQARR303 pKa = 11.84FLSRR307 pKa = 11.84GACC310 pKa = 3.33
MM1 pKa = 7.9RR2 pKa = 11.84ISTAVSWLVVCAGVAALGAVVWTDD26 pKa = 3.16RR27 pKa = 11.84VARR30 pKa = 11.84AVVPRR35 pKa = 11.84AVGAGLLALLVGVTLGRR52 pKa = 11.84LASTRR57 pKa = 11.84TRR59 pKa = 11.84QASTGLGAPRR69 pKa = 11.84VWPVIVIGGVLGSAVLATIGALGGGLVALPIVEE102 pKa = 4.72APLTVEE108 pKa = 4.57ALRR111 pKa = 11.84TTPLVRR117 pKa = 11.84RR118 pKa = 11.84AALAGVVLTHH128 pKa = 6.96ALAGAAGLTTLAHH141 pKa = 7.0TITPGDD147 pKa = 3.35RR148 pKa = 11.84VAATGPAISLAAIAAAGLALAPIGTVPMAGLTVALGALGVGLYY191 pKa = 10.48ARR193 pKa = 11.84APIRR197 pKa = 11.84ALLDD201 pKa = 3.46RR202 pKa = 11.84LGLDD206 pKa = 3.4ARR208 pKa = 11.84VLPARR213 pKa = 11.84SEE215 pKa = 4.16WAHH218 pKa = 6.55DD219 pKa = 3.5AADD222 pKa = 3.45AEE224 pKa = 4.54RR225 pKa = 11.84AHH227 pKa = 7.2RR228 pKa = 11.84RR229 pKa = 11.84AQKK232 pKa = 10.07RR233 pKa = 11.84APPLDD238 pKa = 3.43IGEE241 pKa = 4.37TTSIVVQEE249 pKa = 4.22RR250 pKa = 11.84LDD252 pKa = 3.48GGRR255 pKa = 11.84QLRR258 pKa = 11.84GVYY261 pKa = 10.26EE262 pKa = 4.2DD263 pKa = 3.62FQVFVKK269 pKa = 10.18WDD271 pKa = 3.75VPPDD275 pKa = 3.66CATGEE280 pKa = 4.45TICVRR285 pKa = 11.84IEE287 pKa = 4.23DD288 pKa = 4.44YY289 pKa = 9.47GTNDD293 pKa = 2.76AGQRR297 pKa = 11.84TSGQARR303 pKa = 11.84FLSRR307 pKa = 11.84GACC310 pKa = 3.33
Molecular weight: 31.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
790179 |
36 |
3324 |
312.1 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.728 ± 0.082 | 0.69 ± 0.015 |
9.215 ± 0.059 | 7.564 ± 0.061 |
2.916 ± 0.027 | 8.468 ± 0.049 |
2.009 ± 0.02 | 4.448 ± 0.036 |
1.385 ± 0.026 | 8.485 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.621 ± 0.021 | 2.012 ± 0.036 |
4.962 ± 0.036 | 2.455 ± 0.03 |
6.961 ± 0.056 | 5.545 ± 0.042 |
6.776 ± 0.055 | 9.14 ± 0.048 |
1.188 ± 0.022 | 2.431 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
