
Enterobacteria phage SP (Bacteriophage SP)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Allassoviricetes; Levivirales; Leviviridae; Allolevivirus; Escherichia virus FI
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
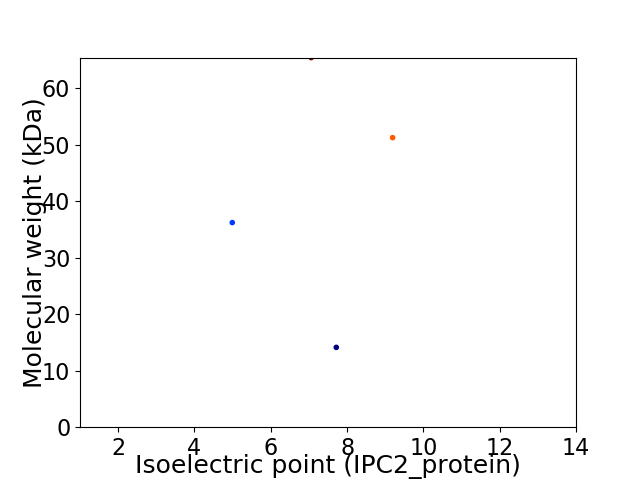
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

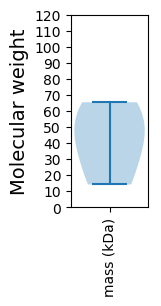
Protein with the lowest isoelectric point:
>sp|P09677|A1_BPSP Minor capsid protein A1 OS=Enterobacteria phage SP OX=12027 PE=2 SV=1
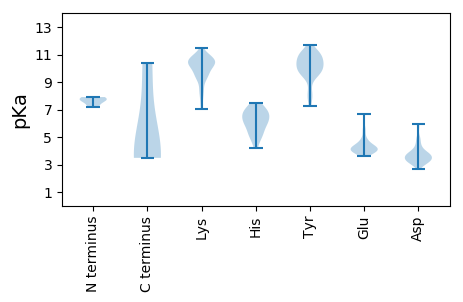
MM1 pKa = 7.71AKK3 pKa = 10.19LNQVTLSKK11 pKa = 10.24IGKK14 pKa = 9.4NGDD17 pKa = 3.22QTLTLTPRR25 pKa = 11.84GVNPTNGVASLSEE38 pKa = 4.3AGAVPALEE46 pKa = 4.02KK47 pKa = 10.58RR48 pKa = 11.84VTVSVAQPSRR58 pKa = 11.84NRR60 pKa = 11.84KK61 pKa = 7.49NFKK64 pKa = 10.37VQIKK68 pKa = 9.11LQNPTACTRR77 pKa = 11.84DD78 pKa = 3.24ACDD81 pKa = 3.43PSVTRR86 pKa = 11.84SAFADD91 pKa = 3.47VTLSFTSYY99 pKa = 9.59STDD102 pKa = 3.16EE103 pKa = 3.89EE104 pKa = 4.2RR105 pKa = 11.84ALIRR109 pKa = 11.84TEE111 pKa = 3.98LAALLADD118 pKa = 4.35PLIVDD123 pKa = 5.92AIDD126 pKa = 3.96NLNPAYY132 pKa = 9.34WAALLVASSGGGDD145 pKa = 3.58NPSDD149 pKa = 3.72PDD151 pKa = 3.77VPVVPDD157 pKa = 3.66VKK159 pKa = 10.85PPDD162 pKa = 3.18GTGRR166 pKa = 11.84YY167 pKa = 7.96KK168 pKa = 10.95CPFACYY174 pKa = 9.56RR175 pKa = 11.84LGSIYY180 pKa = 10.43EE181 pKa = 4.09VGKK184 pKa = 10.57EE185 pKa = 4.09GSPDD189 pKa = 2.68IYY191 pKa = 11.07EE192 pKa = 4.63RR193 pKa = 11.84GDD195 pKa = 3.62EE196 pKa = 4.15VSVTFDD202 pKa = 3.26YY203 pKa = 11.3ALEE206 pKa = 4.6DD207 pKa = 4.09FLGNTNWRR215 pKa = 11.84NWDD218 pKa = 3.54QRR220 pKa = 11.84LSDD223 pKa = 3.69YY224 pKa = 11.14DD225 pKa = 3.03IANRR229 pKa = 11.84RR230 pKa = 11.84RR231 pKa = 11.84CRR233 pKa = 11.84GNGYY237 pKa = 9.75IDD239 pKa = 4.98LDD241 pKa = 3.78ATAMQSDD248 pKa = 4.34DD249 pKa = 3.54FVLSGRR255 pKa = 11.84YY256 pKa = 8.51GVRR259 pKa = 11.84KK260 pKa = 10.15VKK262 pKa = 10.62FPGAFGSIKK271 pKa = 10.58YY272 pKa = 10.02LLNIQGDD279 pKa = 3.36AWLDD283 pKa = 3.65LSEE286 pKa = 4.15VTAYY290 pKa = 10.4RR291 pKa = 11.84SYY293 pKa = 11.7GMVIGFWTDD302 pKa = 2.71SKK304 pKa = 11.46SPQLPTDD311 pKa = 4.28FTQFNSANCPVQTVIIIPSLL331 pKa = 3.49
MM1 pKa = 7.71AKK3 pKa = 10.19LNQVTLSKK11 pKa = 10.24IGKK14 pKa = 9.4NGDD17 pKa = 3.22QTLTLTPRR25 pKa = 11.84GVNPTNGVASLSEE38 pKa = 4.3AGAVPALEE46 pKa = 4.02KK47 pKa = 10.58RR48 pKa = 11.84VTVSVAQPSRR58 pKa = 11.84NRR60 pKa = 11.84KK61 pKa = 7.49NFKK64 pKa = 10.37VQIKK68 pKa = 9.11LQNPTACTRR77 pKa = 11.84DD78 pKa = 3.24ACDD81 pKa = 3.43PSVTRR86 pKa = 11.84SAFADD91 pKa = 3.47VTLSFTSYY99 pKa = 9.59STDD102 pKa = 3.16EE103 pKa = 3.89EE104 pKa = 4.2RR105 pKa = 11.84ALIRR109 pKa = 11.84TEE111 pKa = 3.98LAALLADD118 pKa = 4.35PLIVDD123 pKa = 5.92AIDD126 pKa = 3.96NLNPAYY132 pKa = 9.34WAALLVASSGGGDD145 pKa = 3.58NPSDD149 pKa = 3.72PDD151 pKa = 3.77VPVVPDD157 pKa = 3.66VKK159 pKa = 10.85PPDD162 pKa = 3.18GTGRR166 pKa = 11.84YY167 pKa = 7.96KK168 pKa = 10.95CPFACYY174 pKa = 9.56RR175 pKa = 11.84LGSIYY180 pKa = 10.43EE181 pKa = 4.09VGKK184 pKa = 10.57EE185 pKa = 4.09GSPDD189 pKa = 2.68IYY191 pKa = 11.07EE192 pKa = 4.63RR193 pKa = 11.84GDD195 pKa = 3.62EE196 pKa = 4.15VSVTFDD202 pKa = 3.26YY203 pKa = 11.3ALEE206 pKa = 4.6DD207 pKa = 4.09FLGNTNWRR215 pKa = 11.84NWDD218 pKa = 3.54QRR220 pKa = 11.84LSDD223 pKa = 3.69YY224 pKa = 11.14DD225 pKa = 3.03IANRR229 pKa = 11.84RR230 pKa = 11.84RR231 pKa = 11.84CRR233 pKa = 11.84GNGYY237 pKa = 9.75IDD239 pKa = 4.98LDD241 pKa = 3.78ATAMQSDD248 pKa = 4.34DD249 pKa = 3.54FVLSGRR255 pKa = 11.84YY256 pKa = 8.51GVRR259 pKa = 11.84KK260 pKa = 10.15VKK262 pKa = 10.62FPGAFGSIKK271 pKa = 10.58YY272 pKa = 10.02LLNIQGDD279 pKa = 3.36AWLDD283 pKa = 3.65LSEE286 pKa = 4.15VTAYY290 pKa = 10.4RR291 pKa = 11.84SYY293 pKa = 11.7GMVIGFWTDD302 pKa = 2.71SKK304 pKa = 11.46SPQLPTDD311 pKa = 4.28FTQFNSANCPVQTVIIIPSLL331 pKa = 3.49
Molecular weight: 36.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P09677|A1_BPSP Minor capsid protein A1 OS=Enterobacteria phage SP OX=12027 PE=2 SV=1
MM1 pKa = 7.17PTLPRR6 pKa = 11.84GLRR9 pKa = 11.84FGSNGEE15 pKa = 3.97VLNDD19 pKa = 4.97FEE21 pKa = 5.65ALWFPEE27 pKa = 3.68RR28 pKa = 11.84HH29 pKa = 5.32TVDD32 pKa = 4.7LSNGTCKK39 pKa = 9.99LTGYY43 pKa = 8.13ITNLPGYY50 pKa = 10.65SDD52 pKa = 3.66IFPNKK57 pKa = 9.32GVTAARR63 pKa = 11.84TPYY66 pKa = 10.4RR67 pKa = 11.84STVPVNHH74 pKa = 6.82LGYY77 pKa = 10.5RR78 pKa = 11.84PVTTVEE84 pKa = 4.23YY85 pKa = 10.01IPDD88 pKa = 3.63GTYY91 pKa = 10.56VRR93 pKa = 11.84LDD95 pKa = 3.21GHH97 pKa = 6.06VKK99 pKa = 10.68FEE101 pKa = 4.2GDD103 pKa = 3.64LVNGSVDD110 pKa = 3.31LTNFVISLAAQGGFDD125 pKa = 3.92YY126 pKa = 11.11QSVIGPRR133 pKa = 11.84FSARR137 pKa = 11.84FSAFSTKK144 pKa = 10.58YY145 pKa = 10.48GVLLGEE151 pKa = 4.26GRR153 pKa = 11.84EE154 pKa = 4.09TLKK157 pKa = 11.12YY158 pKa = 10.15LLLVVRR164 pKa = 11.84RR165 pKa = 11.84MRR167 pKa = 11.84EE168 pKa = 3.72GYY170 pKa = 9.2RR171 pKa = 11.84AVRR174 pKa = 11.84RR175 pKa = 11.84GDD177 pKa = 3.48LKK179 pKa = 10.75RR180 pKa = 11.84LRR182 pKa = 11.84NVISTFEE189 pKa = 3.9PSTIKK194 pKa = 10.57GKK196 pKa = 9.62RR197 pKa = 11.84ARR199 pKa = 11.84AEE201 pKa = 3.83FSQTYY206 pKa = 9.68RR207 pKa = 11.84DD208 pKa = 3.74KK209 pKa = 10.98LTGNKK214 pKa = 9.44VEE216 pKa = 4.12VRR218 pKa = 11.84PSEE221 pKa = 4.3GKK223 pKa = 9.22WNSSSASDD231 pKa = 3.0LWLEE235 pKa = 3.89FRR237 pKa = 11.84YY238 pKa = 10.92GLMPLFYY245 pKa = 10.26DD246 pKa = 3.45IQSVMEE252 pKa = 4.32DD253 pKa = 3.16FMRR256 pKa = 11.84VHH258 pKa = 6.76KK259 pKa = 10.46KK260 pKa = 8.56IAKK263 pKa = 8.24IQRR266 pKa = 11.84FSAGHH271 pKa = 6.04GKK273 pKa = 10.43LEE275 pKa = 4.44TVSSRR280 pKa = 11.84FYY282 pKa = 10.62PDD284 pKa = 2.75VHH286 pKa = 6.75FSLEE290 pKa = 4.03VTAVLQRR297 pKa = 11.84RR298 pKa = 11.84HH299 pKa = 5.06RR300 pKa = 11.84WGVIYY305 pKa = 10.45QDD307 pKa = 3.34TGSFATFNNGRR318 pKa = 11.84LVPVKK323 pKa = 10.05DD324 pKa = 3.07WKK326 pKa = 9.34TAAFALLNPAEE337 pKa = 4.61VAWEE341 pKa = 3.92VTPYY345 pKa = 11.07SFVVDD350 pKa = 3.11WFVNVGDD357 pKa = 4.51MLEE360 pKa = 4.16QMGQLYY366 pKa = 9.74RR367 pKa = 11.84HH368 pKa = 6.16VDD370 pKa = 3.45VVDD373 pKa = 3.84GFDD376 pKa = 4.27RR377 pKa = 11.84KK378 pKa = 10.39DD379 pKa = 3.13IKK381 pKa = 10.88LKK383 pKa = 9.86SVSVRR388 pKa = 11.84VLTNDD393 pKa = 3.15VAHH396 pKa = 6.1VASFQLRR403 pKa = 11.84QAKK406 pKa = 9.53LLHH409 pKa = 6.21SYY411 pKa = 9.86YY412 pKa = 10.93SRR414 pKa = 11.84VHH416 pKa = 5.56TVAFPQISPQLDD428 pKa = 3.44TEE430 pKa = 4.2IRR432 pKa = 11.84SVKK435 pKa = 10.36HH436 pKa = 5.94VIDD439 pKa = 4.9SIALLTQRR447 pKa = 11.84VKK449 pKa = 10.89RR450 pKa = 3.95
MM1 pKa = 7.17PTLPRR6 pKa = 11.84GLRR9 pKa = 11.84FGSNGEE15 pKa = 3.97VLNDD19 pKa = 4.97FEE21 pKa = 5.65ALWFPEE27 pKa = 3.68RR28 pKa = 11.84HH29 pKa = 5.32TVDD32 pKa = 4.7LSNGTCKK39 pKa = 9.99LTGYY43 pKa = 8.13ITNLPGYY50 pKa = 10.65SDD52 pKa = 3.66IFPNKK57 pKa = 9.32GVTAARR63 pKa = 11.84TPYY66 pKa = 10.4RR67 pKa = 11.84STVPVNHH74 pKa = 6.82LGYY77 pKa = 10.5RR78 pKa = 11.84PVTTVEE84 pKa = 4.23YY85 pKa = 10.01IPDD88 pKa = 3.63GTYY91 pKa = 10.56VRR93 pKa = 11.84LDD95 pKa = 3.21GHH97 pKa = 6.06VKK99 pKa = 10.68FEE101 pKa = 4.2GDD103 pKa = 3.64LVNGSVDD110 pKa = 3.31LTNFVISLAAQGGFDD125 pKa = 3.92YY126 pKa = 11.11QSVIGPRR133 pKa = 11.84FSARR137 pKa = 11.84FSAFSTKK144 pKa = 10.58YY145 pKa = 10.48GVLLGEE151 pKa = 4.26GRR153 pKa = 11.84EE154 pKa = 4.09TLKK157 pKa = 11.12YY158 pKa = 10.15LLLVVRR164 pKa = 11.84RR165 pKa = 11.84MRR167 pKa = 11.84EE168 pKa = 3.72GYY170 pKa = 9.2RR171 pKa = 11.84AVRR174 pKa = 11.84RR175 pKa = 11.84GDD177 pKa = 3.48LKK179 pKa = 10.75RR180 pKa = 11.84LRR182 pKa = 11.84NVISTFEE189 pKa = 3.9PSTIKK194 pKa = 10.57GKK196 pKa = 9.62RR197 pKa = 11.84ARR199 pKa = 11.84AEE201 pKa = 3.83FSQTYY206 pKa = 9.68RR207 pKa = 11.84DD208 pKa = 3.74KK209 pKa = 10.98LTGNKK214 pKa = 9.44VEE216 pKa = 4.12VRR218 pKa = 11.84PSEE221 pKa = 4.3GKK223 pKa = 9.22WNSSSASDD231 pKa = 3.0LWLEE235 pKa = 3.89FRR237 pKa = 11.84YY238 pKa = 10.92GLMPLFYY245 pKa = 10.26DD246 pKa = 3.45IQSVMEE252 pKa = 4.32DD253 pKa = 3.16FMRR256 pKa = 11.84VHH258 pKa = 6.76KK259 pKa = 10.46KK260 pKa = 8.56IAKK263 pKa = 8.24IQRR266 pKa = 11.84FSAGHH271 pKa = 6.04GKK273 pKa = 10.43LEE275 pKa = 4.44TVSSRR280 pKa = 11.84FYY282 pKa = 10.62PDD284 pKa = 2.75VHH286 pKa = 6.75FSLEE290 pKa = 4.03VTAVLQRR297 pKa = 11.84RR298 pKa = 11.84HH299 pKa = 5.06RR300 pKa = 11.84WGVIYY305 pKa = 10.45QDD307 pKa = 3.34TGSFATFNNGRR318 pKa = 11.84LVPVKK323 pKa = 10.05DD324 pKa = 3.07WKK326 pKa = 9.34TAAFALLNPAEE337 pKa = 4.61VAWEE341 pKa = 3.92VTPYY345 pKa = 11.07SFVVDD350 pKa = 3.11WFVNVGDD357 pKa = 4.51MLEE360 pKa = 4.16QMGQLYY366 pKa = 9.74RR367 pKa = 11.84HH368 pKa = 6.16VDD370 pKa = 3.45VVDD373 pKa = 3.84GFDD376 pKa = 4.27RR377 pKa = 11.84KK378 pKa = 10.39DD379 pKa = 3.13IKK381 pKa = 10.88LKK383 pKa = 9.86SVSVRR388 pKa = 11.84VLTNDD393 pKa = 3.15VAHH396 pKa = 6.1VASFQLRR403 pKa = 11.84QAKK406 pKa = 9.53LLHH409 pKa = 6.21SYY411 pKa = 9.86YY412 pKa = 10.93SRR414 pKa = 11.84VHH416 pKa = 5.56TVAFPQISPQLDD428 pKa = 3.44TEE430 pKa = 4.2IRR432 pKa = 11.84SVKK435 pKa = 10.36HH436 pKa = 5.94VIDD439 pKa = 4.9SIALLTQRR447 pKa = 11.84VKK449 pKa = 10.89RR450 pKa = 3.95
Molecular weight: 51.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1489 |
132 |
576 |
372.3 |
41.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.455 ± 0.726 | 1.276 ± 0.386 |
6.985 ± 0.589 | 4.634 ± 0.516 |
4.365 ± 0.492 | 6.514 ± 0.376 |
1.41 ± 0.5 | 4.835 ± 0.682 |
4.835 ± 0.169 | 9.604 ± 0.449 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.209 ± 0.128 | 4.231 ± 0.481 |
5.037 ± 0.404 | 2.821 ± 0.326 |
7.79 ± 0.558 | 7.052 ± 0.368 |
6.38 ± 0.442 | 8.328 ± 0.941 |
1.545 ± 0.196 | 3.694 ± 0.263 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
