
Jodiemicrovirus-1
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Gokushovirinae; unclassified Gokushovirinae
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
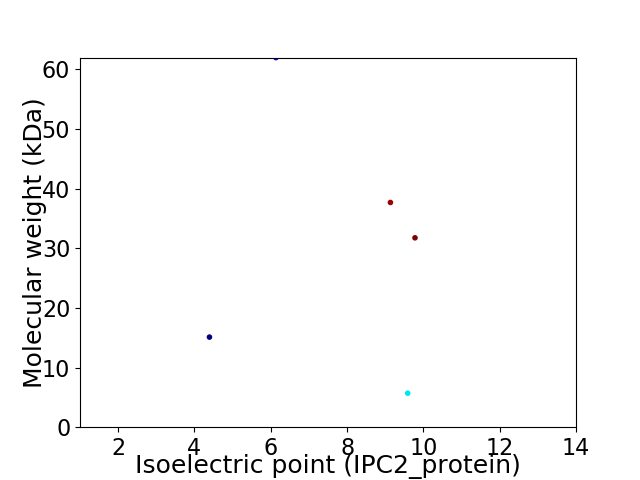
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
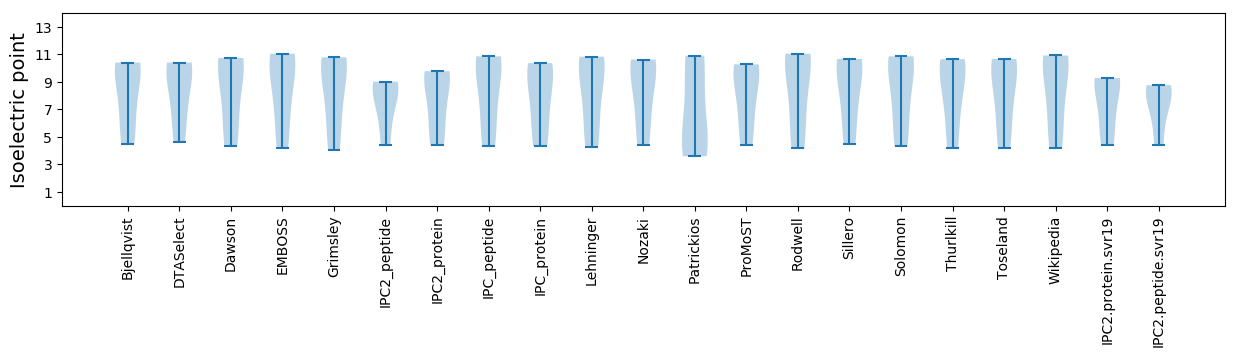
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C0PWM8|A0A5C0PWM8_9VIRU Major capsid protein OS=Jodiemicrovirus-1 OX=2603850 PE=3 SV=1
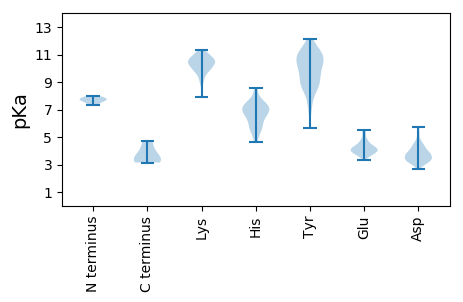
MM1 pKa = 7.31FKK3 pKa = 10.52KK4 pKa = 9.94IRR6 pKa = 11.84YY7 pKa = 5.66VTIHH11 pKa = 6.56NYY13 pKa = 10.47KK14 pKa = 10.66EE15 pKa = 3.87MDD17 pKa = 3.33SSFEE21 pKa = 4.04YY22 pKa = 11.02VNPTKK27 pKa = 10.82VVDD30 pKa = 3.88DD31 pKa = 3.9VGYY34 pKa = 11.08LNLYY38 pKa = 8.61QYY40 pKa = 11.89AMRR43 pKa = 11.84ALKK46 pKa = 10.57GYY48 pKa = 9.48IVPISSLKK56 pKa = 9.37YY57 pKa = 10.48DD58 pKa = 3.83EE59 pKa = 5.48DD60 pKa = 4.47VSVDD64 pKa = 4.59DD65 pKa = 3.66IHH67 pKa = 8.52KK68 pKa = 10.56SASQSEE74 pKa = 4.4NQKK77 pKa = 10.5SEE79 pKa = 4.27AKK81 pKa = 10.56DD82 pKa = 3.66EE83 pKa = 4.46LNQADD88 pKa = 5.27DD89 pKa = 3.87EE90 pKa = 4.83LEE92 pKa = 4.28LYY94 pKa = 8.5EE95 pKa = 4.55AAMDD99 pKa = 4.23AGSIHH104 pKa = 7.06HH105 pKa = 7.44DD106 pKa = 2.95IDD108 pKa = 3.66PTVEE112 pKa = 4.09NEE114 pKa = 4.18KK115 pKa = 9.92STDD118 pKa = 3.73LPDD121 pKa = 3.92KK122 pKa = 10.97SEE124 pKa = 4.24PASSEE129 pKa = 3.94ATKK132 pKa = 10.73DD133 pKa = 3.21
MM1 pKa = 7.31FKK3 pKa = 10.52KK4 pKa = 9.94IRR6 pKa = 11.84YY7 pKa = 5.66VTIHH11 pKa = 6.56NYY13 pKa = 10.47KK14 pKa = 10.66EE15 pKa = 3.87MDD17 pKa = 3.33SSFEE21 pKa = 4.04YY22 pKa = 11.02VNPTKK27 pKa = 10.82VVDD30 pKa = 3.88DD31 pKa = 3.9VGYY34 pKa = 11.08LNLYY38 pKa = 8.61QYY40 pKa = 11.89AMRR43 pKa = 11.84ALKK46 pKa = 10.57GYY48 pKa = 9.48IVPISSLKK56 pKa = 9.37YY57 pKa = 10.48DD58 pKa = 3.83EE59 pKa = 5.48DD60 pKa = 4.47VSVDD64 pKa = 4.59DD65 pKa = 3.66IHH67 pKa = 8.52KK68 pKa = 10.56SASQSEE74 pKa = 4.4NQKK77 pKa = 10.5SEE79 pKa = 4.27AKK81 pKa = 10.56DD82 pKa = 3.66EE83 pKa = 4.46LNQADD88 pKa = 5.27DD89 pKa = 3.87EE90 pKa = 4.83LEE92 pKa = 4.28LYY94 pKa = 8.5EE95 pKa = 4.55AAMDD99 pKa = 4.23AGSIHH104 pKa = 7.06HH105 pKa = 7.44DD106 pKa = 2.95IDD108 pKa = 3.66PTVEE112 pKa = 4.09NEE114 pKa = 4.18KK115 pKa = 9.92STDD118 pKa = 3.73LPDD121 pKa = 3.92KK122 pKa = 10.97SEE124 pKa = 4.24PASSEE129 pKa = 3.94ATKK132 pKa = 10.73DD133 pKa = 3.21
Molecular weight: 15.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C0PZD8|A0A5C0PZD8_9VIRU Minor capsid protein OS=Jodiemicrovirus-1 OX=2603850 PE=4 SV=1
MM1 pKa = 7.73LGPLSRR7 pKa = 11.84FLISAAPAVVKK18 pKa = 10.47LFTRR22 pKa = 11.84SKK24 pKa = 9.18PRR26 pKa = 11.84PPGRR30 pKa = 11.84VNVPPARR37 pKa = 11.84PIRR40 pKa = 11.84SIDD43 pKa = 3.53DD44 pKa = 4.15FYY46 pKa = 12.15SKK48 pKa = 11.18LNASKK53 pKa = 10.81SPSWFNRR60 pKa = 11.84LLKK63 pKa = 10.77LGAAGGAAGALGSLSPSSTGDD84 pKa = 3.41LPSGVYY90 pKa = 10.51SNPSRR95 pKa = 11.84FGYY98 pKa = 9.69FDD100 pKa = 3.59KK101 pKa = 11.26VNKK104 pKa = 10.26LSSQYY109 pKa = 9.19QNRR112 pKa = 11.84FSGFANNLDD121 pKa = 4.29TNASTPSVSGSKK133 pKa = 10.21LLSGIPGNLLSLAGGIGTGLFSRR156 pKa = 11.84YY157 pKa = 8.98LDD159 pKa = 3.24RR160 pKa = 11.84KK161 pKa = 10.44SINQSNAIQQAAEE174 pKa = 3.61DD175 pKa = 4.15RR176 pKa = 11.84AWKK179 pKa = 10.58RR180 pKa = 11.84EE181 pKa = 3.75TEE183 pKa = 4.32YY184 pKa = 11.38NHH186 pKa = 7.12PAPQMARR193 pKa = 11.84FRR195 pKa = 11.84EE196 pKa = 4.37AGLNPALMYY205 pKa = 10.7SSSLSSASGTTHH217 pKa = 7.13APLTSRR223 pKa = 11.84SSYY226 pKa = 8.45YY227 pKa = 9.29QDD229 pKa = 5.17RR230 pKa = 11.84IASYY234 pKa = 10.81LSLANMHH241 pKa = 6.36AQNKK245 pKa = 9.61LLDD248 pKa = 3.92AQARR252 pKa = 11.84SVNADD257 pKa = 3.07VFGKK261 pKa = 10.52EE262 pKa = 3.85KK263 pKa = 10.41INRR266 pKa = 11.84VLGDD270 pKa = 3.66YY271 pKa = 10.72GFSTDD276 pKa = 3.82PTWLRR281 pKa = 11.84SIFRR285 pKa = 11.84FIDD288 pKa = 3.22YY289 pKa = 10.65RR290 pKa = 11.84NNN292 pKa = 3.15
MM1 pKa = 7.73LGPLSRR7 pKa = 11.84FLISAAPAVVKK18 pKa = 10.47LFTRR22 pKa = 11.84SKK24 pKa = 9.18PRR26 pKa = 11.84PPGRR30 pKa = 11.84VNVPPARR37 pKa = 11.84PIRR40 pKa = 11.84SIDD43 pKa = 3.53DD44 pKa = 4.15FYY46 pKa = 12.15SKK48 pKa = 11.18LNASKK53 pKa = 10.81SPSWFNRR60 pKa = 11.84LLKK63 pKa = 10.77LGAAGGAAGALGSLSPSSTGDD84 pKa = 3.41LPSGVYY90 pKa = 10.51SNPSRR95 pKa = 11.84FGYY98 pKa = 9.69FDD100 pKa = 3.59KK101 pKa = 11.26VNKK104 pKa = 10.26LSSQYY109 pKa = 9.19QNRR112 pKa = 11.84FSGFANNLDD121 pKa = 4.29TNASTPSVSGSKK133 pKa = 10.21LLSGIPGNLLSLAGGIGTGLFSRR156 pKa = 11.84YY157 pKa = 8.98LDD159 pKa = 3.24RR160 pKa = 11.84KK161 pKa = 10.44SINQSNAIQQAAEE174 pKa = 3.61DD175 pKa = 4.15RR176 pKa = 11.84AWKK179 pKa = 10.58RR180 pKa = 11.84EE181 pKa = 3.75TEE183 pKa = 4.32YY184 pKa = 11.38NHH186 pKa = 7.12PAPQMARR193 pKa = 11.84FRR195 pKa = 11.84EE196 pKa = 4.37AGLNPALMYY205 pKa = 10.7SSSLSSASGTTHH217 pKa = 7.13APLTSRR223 pKa = 11.84SSYY226 pKa = 8.45YY227 pKa = 9.29QDD229 pKa = 5.17RR230 pKa = 11.84IASYY234 pKa = 10.81LSLANMHH241 pKa = 6.36AQNKK245 pKa = 9.61LLDD248 pKa = 3.92AQARR252 pKa = 11.84SVNADD257 pKa = 3.07VFGKK261 pKa = 10.52EE262 pKa = 3.85KK263 pKa = 10.41INRR266 pKa = 11.84VLGDD270 pKa = 3.66YY271 pKa = 10.72GFSTDD276 pKa = 3.82PTWLRR281 pKa = 11.84SIFRR285 pKa = 11.84FIDD288 pKa = 3.22YY289 pKa = 10.65RR290 pKa = 11.84NNN292 pKa = 3.15
Molecular weight: 31.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1343 |
49 |
552 |
268.6 |
30.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.85 ± 1.074 | 1.191 ± 0.637 |
6.031 ± 0.828 | 4.393 ± 0.902 |
5.733 ± 0.87 | 5.659 ± 1.077 |
2.681 ± 0.589 | 4.914 ± 0.373 |
6.329 ± 1.661 | 8.786 ± 0.724 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.936 ± 0.254 | 4.84 ± 0.607 |
4.84 ± 0.905 | 2.978 ± 0.076 |
6.478 ± 0.678 | 10.276 ± 1.081 |
4.989 ± 0.701 | 5.436 ± 0.708 |
1.117 ± 0.214 | 4.542 ± 0.619 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
