
Populus trichocarpa (Western balsam poplar) (Populus balsamifera subsp. trichocarpa)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
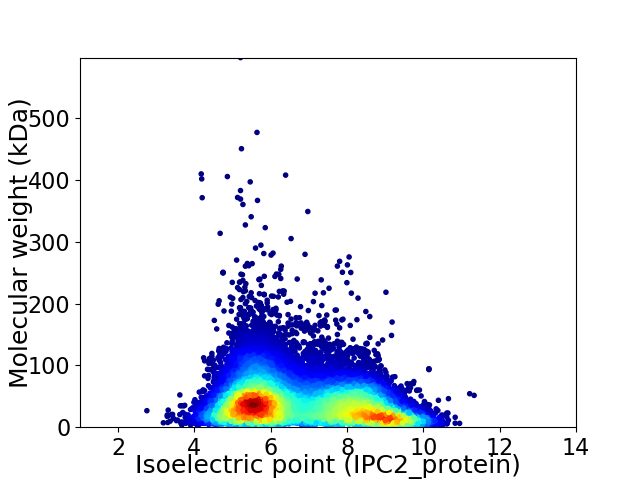
Virtual 2D-PAGE plot for 53336 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
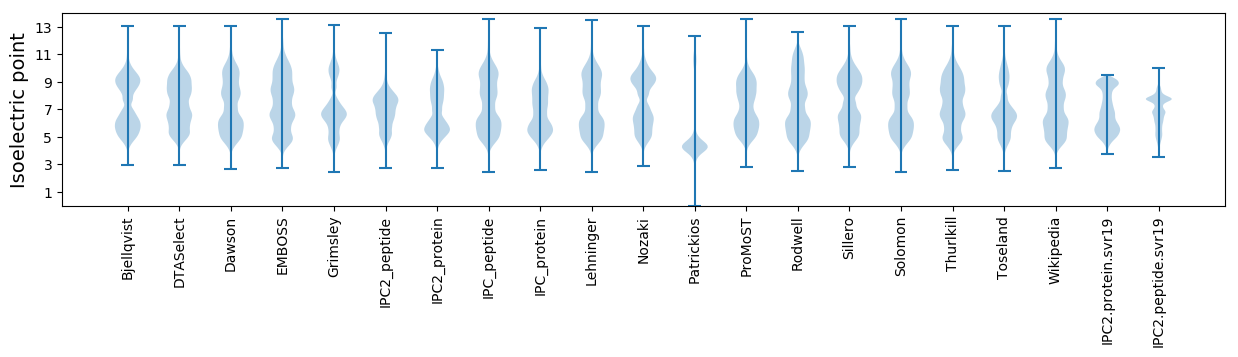
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B9IC68|B9IC68_POPTR Uncharacterized protein OS=Populus trichocarpa OX=3694 GN=POPTR_014G076300 PE=4 SV=2
MM1 pKa = 7.81DD2 pKa = 4.19RR3 pKa = 11.84VKK5 pKa = 11.18NSVDD9 pKa = 3.21VSPFLLVEE17 pKa = 4.01AAGDD21 pKa = 4.1SEE23 pKa = 4.56VDD25 pKa = 3.22SDD27 pKa = 4.01PTMSTIDD34 pKa = 4.97LVDD37 pKa = 5.01DD38 pKa = 5.26DD39 pKa = 6.28DD40 pKa = 5.25DD41 pKa = 6.6AEE43 pKa = 4.43SCSCDD48 pKa = 3.46TSDD51 pKa = 4.3HH52 pKa = 6.46SCVSDD57 pKa = 4.11IIKK60 pKa = 9.99GACSEE65 pKa = 4.29VEE67 pKa = 4.11AYY69 pKa = 9.01QVNYY73 pKa = 10.83NVVDD77 pKa = 4.31DD78 pKa = 5.72DD79 pKa = 4.69DD80 pKa = 6.63DD81 pKa = 4.44EE82 pKa = 4.94EE83 pKa = 4.63EE84 pKa = 4.27EE85 pKa = 4.36EE86 pKa = 4.58EE87 pKa = 4.39GVEE90 pKa = 4.54VCQSWVHH97 pKa = 5.62HH98 pKa = 5.03VHH100 pKa = 6.06VGLPVKK106 pKa = 10.37QKK108 pKa = 10.32SCVSVDD114 pKa = 3.51SSNEE118 pKa = 3.73SMNEE122 pKa = 3.75KK123 pKa = 10.48EE124 pKa = 4.17KK125 pKa = 11.35DD126 pKa = 3.38RR127 pKa = 11.84LFWEE131 pKa = 4.71ACLASS136 pKa = 3.65
MM1 pKa = 7.81DD2 pKa = 4.19RR3 pKa = 11.84VKK5 pKa = 11.18NSVDD9 pKa = 3.21VSPFLLVEE17 pKa = 4.01AAGDD21 pKa = 4.1SEE23 pKa = 4.56VDD25 pKa = 3.22SDD27 pKa = 4.01PTMSTIDD34 pKa = 4.97LVDD37 pKa = 5.01DD38 pKa = 5.26DD39 pKa = 6.28DD40 pKa = 5.25DD41 pKa = 6.6AEE43 pKa = 4.43SCSCDD48 pKa = 3.46TSDD51 pKa = 4.3HH52 pKa = 6.46SCVSDD57 pKa = 4.11IIKK60 pKa = 9.99GACSEE65 pKa = 4.29VEE67 pKa = 4.11AYY69 pKa = 9.01QVNYY73 pKa = 10.83NVVDD77 pKa = 4.31DD78 pKa = 5.72DD79 pKa = 4.69DD80 pKa = 6.63DD81 pKa = 4.44EE82 pKa = 4.94EE83 pKa = 4.63EE84 pKa = 4.27EE85 pKa = 4.36EE86 pKa = 4.58EE87 pKa = 4.39GVEE90 pKa = 4.54VCQSWVHH97 pKa = 5.62HH98 pKa = 5.03VHH100 pKa = 6.06VGLPVKK106 pKa = 10.37QKK108 pKa = 10.32SCVSVDD114 pKa = 3.51SSNEE118 pKa = 3.73SMNEE122 pKa = 3.75KK123 pKa = 10.48EE124 pKa = 4.17KK125 pKa = 11.35DD126 pKa = 3.38RR127 pKa = 11.84LFWEE131 pKa = 4.71ACLASS136 pKa = 3.65
Molecular weight: 14.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N7EZH0|A0A3N7EZH0_POPTR Uncharacterized protein OS=Populus trichocarpa OX=3694 GN=POPTR_005G126600 PE=4 SV=1
MM1 pKa = 7.49SLIIINLHH9 pKa = 5.8HH10 pKa = 6.8LHH12 pKa = 6.68LNHH15 pKa = 6.89HH16 pKa = 6.29LHH18 pKa = 7.33HH19 pKa = 6.72ITTHH23 pKa = 6.15HH24 pKa = 6.98LLHH27 pKa = 6.29QKK29 pKa = 10.02SPPLHH34 pKa = 7.08LITTHH39 pKa = 5.92PHH41 pKa = 5.95LLQRR45 pKa = 11.84SPHH48 pKa = 6.02HH49 pKa = 6.2LHH51 pKa = 5.79TTTHH55 pKa = 6.85PLLLQRR61 pKa = 11.84SLPHH65 pKa = 6.33LHH67 pKa = 5.85TTTHH71 pKa = 6.87PLLHH75 pKa = 6.42LRR77 pKa = 11.84SLHH80 pKa = 6.1HH81 pKa = 6.53LHH83 pKa = 5.97TTTHH87 pKa = 6.85PLLLQRR93 pKa = 11.84SLPHH97 pKa = 6.33LHH99 pKa = 5.69TTTHH103 pKa = 6.43ILLHH107 pKa = 6.08LRR109 pKa = 11.84SLHH112 pKa = 6.1HH113 pKa = 6.53LHH115 pKa = 5.99TTTHH119 pKa = 5.99PHH121 pKa = 5.44LHH123 pKa = 5.46QRR125 pKa = 11.84SLPLHH130 pKa = 6.21HH131 pKa = 6.85TTTHH135 pKa = 6.62PLLLQRR141 pKa = 11.84SLPHH145 pKa = 6.33LHH147 pKa = 5.85TTTHH151 pKa = 6.85PLLLQRR157 pKa = 11.84SLHH160 pKa = 5.99HH161 pKa = 6.36LHH163 pKa = 5.84TTTHH167 pKa = 6.44ILLHH171 pKa = 6.08LRR173 pKa = 11.84SLHH176 pKa = 6.1HH177 pKa = 6.53LHH179 pKa = 5.99TTTHH183 pKa = 5.99PHH185 pKa = 5.44LHH187 pKa = 5.46QRR189 pKa = 11.84SLPLHH194 pKa = 6.21HH195 pKa = 6.85TTTHH199 pKa = 6.62PLLLQRR205 pKa = 11.84SPHH208 pKa = 6.06HH209 pKa = 6.11LHH211 pKa = 5.68TTTHH215 pKa = 6.86LLLHH219 pKa = 6.38LRR221 pKa = 11.84SLPLHH226 pKa = 6.24HH227 pKa = 6.89TTTHH231 pKa = 6.25HH232 pKa = 7.32LLLQRR237 pKa = 11.84SRR239 pKa = 11.84HH240 pKa = 5.42HH241 pKa = 6.35LHH243 pKa = 5.77TTTHH247 pKa = 6.87PLLHH251 pKa = 6.3LRR253 pKa = 11.84SLLLHH258 pKa = 6.35HH259 pKa = 6.69TTTHH263 pKa = 5.8PHH265 pKa = 5.72HH266 pKa = 7.12LLRR269 pKa = 11.84SLHH272 pKa = 6.24HH273 pKa = 6.5LHH275 pKa = 5.66TTIHH279 pKa = 6.73PLLHH283 pKa = 5.94QRR285 pKa = 11.84SLPHH289 pKa = 6.28LHH291 pKa = 5.85TTTHH295 pKa = 6.47HH296 pKa = 7.13LLLQRR301 pKa = 11.84NLHH304 pKa = 6.06HH305 pKa = 6.9LHH307 pKa = 5.74TTTHH311 pKa = 6.68LLSHH315 pKa = 6.7LRR317 pKa = 11.84SIPLHH322 pKa = 5.87HH323 pKa = 6.8TTTHH327 pKa = 5.44PHH329 pKa = 6.38LLRR332 pKa = 11.84RR333 pKa = 11.84SLHH336 pKa = 5.97HH337 pKa = 6.52LHH339 pKa = 5.94TTTHH343 pKa = 7.32LLPLQRR349 pKa = 11.84SLPLHH354 pKa = 7.09HH355 pKa = 6.57ITIHH359 pKa = 5.28PHH361 pKa = 5.26LLRR364 pKa = 11.84RR365 pKa = 11.84SLHH368 pKa = 5.97HH369 pKa = 6.52LHH371 pKa = 5.96TTTHH375 pKa = 6.29LHH377 pKa = 6.01PLRR380 pKa = 11.84RR381 pKa = 11.84NLPLHH386 pKa = 6.03HH387 pKa = 6.66TTTHH391 pKa = 6.78LLLLQRR397 pKa = 11.84SLPHH401 pKa = 6.33LHH403 pKa = 5.86TTTHH407 pKa = 6.56LPLLHH412 pKa = 6.45HH413 pKa = 6.42HH414 pKa = 6.52HH415 pKa = 6.92HH416 pKa = 6.77LLHH419 pKa = 6.15TCMHH423 pKa = 6.79HH424 pKa = 6.88LHH426 pKa = 6.44HH427 pKa = 6.97QNTTKK432 pKa = 10.24MLL434 pKa = 3.99
MM1 pKa = 7.49SLIIINLHH9 pKa = 5.8HH10 pKa = 6.8LHH12 pKa = 6.68LNHH15 pKa = 6.89HH16 pKa = 6.29LHH18 pKa = 7.33HH19 pKa = 6.72ITTHH23 pKa = 6.15HH24 pKa = 6.98LLHH27 pKa = 6.29QKK29 pKa = 10.02SPPLHH34 pKa = 7.08LITTHH39 pKa = 5.92PHH41 pKa = 5.95LLQRR45 pKa = 11.84SPHH48 pKa = 6.02HH49 pKa = 6.2LHH51 pKa = 5.79TTTHH55 pKa = 6.85PLLLQRR61 pKa = 11.84SLPHH65 pKa = 6.33LHH67 pKa = 5.85TTTHH71 pKa = 6.87PLLHH75 pKa = 6.42LRR77 pKa = 11.84SLHH80 pKa = 6.1HH81 pKa = 6.53LHH83 pKa = 5.97TTTHH87 pKa = 6.85PLLLQRR93 pKa = 11.84SLPHH97 pKa = 6.33LHH99 pKa = 5.69TTTHH103 pKa = 6.43ILLHH107 pKa = 6.08LRR109 pKa = 11.84SLHH112 pKa = 6.1HH113 pKa = 6.53LHH115 pKa = 5.99TTTHH119 pKa = 5.99PHH121 pKa = 5.44LHH123 pKa = 5.46QRR125 pKa = 11.84SLPLHH130 pKa = 6.21HH131 pKa = 6.85TTTHH135 pKa = 6.62PLLLQRR141 pKa = 11.84SLPHH145 pKa = 6.33LHH147 pKa = 5.85TTTHH151 pKa = 6.85PLLLQRR157 pKa = 11.84SLHH160 pKa = 5.99HH161 pKa = 6.36LHH163 pKa = 5.84TTTHH167 pKa = 6.44ILLHH171 pKa = 6.08LRR173 pKa = 11.84SLHH176 pKa = 6.1HH177 pKa = 6.53LHH179 pKa = 5.99TTTHH183 pKa = 5.99PHH185 pKa = 5.44LHH187 pKa = 5.46QRR189 pKa = 11.84SLPLHH194 pKa = 6.21HH195 pKa = 6.85TTTHH199 pKa = 6.62PLLLQRR205 pKa = 11.84SPHH208 pKa = 6.06HH209 pKa = 6.11LHH211 pKa = 5.68TTTHH215 pKa = 6.86LLLHH219 pKa = 6.38LRR221 pKa = 11.84SLPLHH226 pKa = 6.24HH227 pKa = 6.89TTTHH231 pKa = 6.25HH232 pKa = 7.32LLLQRR237 pKa = 11.84SRR239 pKa = 11.84HH240 pKa = 5.42HH241 pKa = 6.35LHH243 pKa = 5.77TTTHH247 pKa = 6.87PLLHH251 pKa = 6.3LRR253 pKa = 11.84SLLLHH258 pKa = 6.35HH259 pKa = 6.69TTTHH263 pKa = 5.8PHH265 pKa = 5.72HH266 pKa = 7.12LLRR269 pKa = 11.84SLHH272 pKa = 6.24HH273 pKa = 6.5LHH275 pKa = 5.66TTIHH279 pKa = 6.73PLLHH283 pKa = 5.94QRR285 pKa = 11.84SLPHH289 pKa = 6.28LHH291 pKa = 5.85TTTHH295 pKa = 6.47HH296 pKa = 7.13LLLQRR301 pKa = 11.84NLHH304 pKa = 6.06HH305 pKa = 6.9LHH307 pKa = 5.74TTTHH311 pKa = 6.68LLSHH315 pKa = 6.7LRR317 pKa = 11.84SIPLHH322 pKa = 5.87HH323 pKa = 6.8TTTHH327 pKa = 5.44PHH329 pKa = 6.38LLRR332 pKa = 11.84RR333 pKa = 11.84SLHH336 pKa = 5.97HH337 pKa = 6.52LHH339 pKa = 5.94TTTHH343 pKa = 7.32LLPLQRR349 pKa = 11.84SLPLHH354 pKa = 7.09HH355 pKa = 6.57ITIHH359 pKa = 5.28PHH361 pKa = 5.26LLRR364 pKa = 11.84RR365 pKa = 11.84SLHH368 pKa = 5.97HH369 pKa = 6.52LHH371 pKa = 5.96TTTHH375 pKa = 6.29LHH377 pKa = 6.01PLRR380 pKa = 11.84RR381 pKa = 11.84NLPLHH386 pKa = 6.03HH387 pKa = 6.66TTTHH391 pKa = 6.78LLLLQRR397 pKa = 11.84SLPHH401 pKa = 6.33LHH403 pKa = 5.86TTTHH407 pKa = 6.56LPLLHH412 pKa = 6.45HH413 pKa = 6.42HH414 pKa = 6.52HH415 pKa = 6.92HH416 pKa = 6.77LLHH419 pKa = 6.15TCMHH423 pKa = 6.79HH424 pKa = 6.88LHH426 pKa = 6.44HH427 pKa = 6.97QNTTKK432 pKa = 10.24MLL434 pKa = 3.99
Molecular weight: 51.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
21108701 |
27 |
5451 |
395.8 |
44.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.522 ± 0.01 | 1.914 ± 0.006 |
5.26 ± 0.008 | 6.492 ± 0.013 |
4.225 ± 0.006 | 6.556 ± 0.01 |
2.395 ± 0.004 | 5.325 ± 0.008 |
6.153 ± 0.009 | 9.837 ± 0.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.453 ± 0.004 | 4.578 ± 0.007 |
4.876 ± 0.009 | 3.8 ± 0.009 |
5.205 ± 0.009 | 9.173 ± 0.012 |
4.864 ± 0.006 | 6.366 ± 0.008 |
1.267 ± 0.004 | 2.739 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
