
Lebombo virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
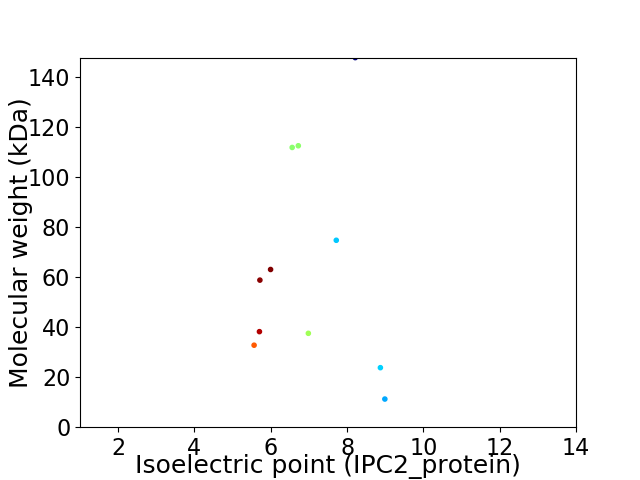
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5QLZ7|W5QLZ7_9REOV RNA-directed RNA polymerase OS=Lebombo virus OX=40057 PE=3 SV=1
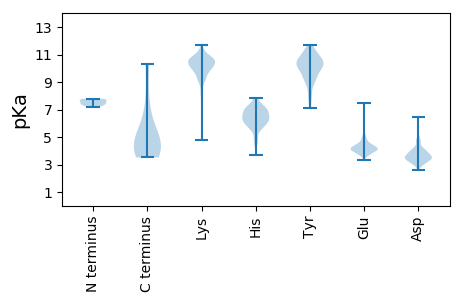
MM1 pKa = 7.29AQEE4 pKa = 3.93QVQVKK9 pKa = 10.12RR10 pKa = 11.84KK11 pKa = 6.29FTRR14 pKa = 11.84TICVLDD20 pKa = 4.26PGAKK24 pKa = 8.56TFCGKK29 pKa = 8.83VAKK32 pKa = 10.7LEE34 pKa = 4.23GKK36 pKa = 8.31TYY38 pKa = 10.32FVIKK42 pKa = 10.24IGRR45 pKa = 11.84TTQVGVSNTPVPKK58 pKa = 10.38CYY60 pKa = 10.56VLDD63 pKa = 3.72VAGPGSYY70 pKa = 9.06RR71 pKa = 11.84VQDD74 pKa = 3.8GQDD77 pKa = 3.18TVSVILSEE85 pKa = 4.49SGFEE89 pKa = 4.01ATGEE93 pKa = 3.84RR94 pKa = 11.84WEE96 pKa = 4.01EE97 pKa = 3.52WRR99 pKa = 11.84FEE101 pKa = 4.21VLSATPIVARR111 pKa = 11.84MAVGEE116 pKa = 4.59DD117 pKa = 3.48EE118 pKa = 4.68FDD120 pKa = 4.63AEE122 pKa = 4.27IKK124 pKa = 10.1YY125 pKa = 10.88SKK127 pKa = 10.81GLGSVPPYY135 pKa = 10.79AKK137 pKa = 10.62NEE139 pKa = 3.91ADD141 pKa = 3.6RR142 pKa = 11.84RR143 pKa = 11.84MMPTLPGVKK152 pKa = 9.91ALDD155 pKa = 3.33MPIRR159 pKa = 11.84DD160 pKa = 3.98FRR162 pKa = 11.84QHH164 pKa = 5.8ARR166 pKa = 11.84EE167 pKa = 3.95HH168 pKa = 6.32RR169 pKa = 11.84DD170 pKa = 3.21EE171 pKa = 4.82VRR173 pKa = 11.84AQLDD177 pKa = 3.37RR178 pKa = 11.84AARR181 pKa = 11.84RR182 pKa = 11.84LDD184 pKa = 3.68PSSSRR189 pKa = 11.84LLGASVGLSEE199 pKa = 5.38EE200 pKa = 4.09IPIEE204 pKa = 4.22SVLPQAKK211 pKa = 9.63RR212 pKa = 11.84EE213 pKa = 4.1PVVFASASRR222 pKa = 11.84EE223 pKa = 3.81LDD225 pKa = 2.96DD226 pKa = 4.87WGFDD230 pKa = 3.73MQPKK234 pKa = 9.62TSGDD238 pKa = 3.27WGAKK242 pKa = 7.51TPIPEE247 pKa = 4.15VEE249 pKa = 4.07SDD251 pKa = 3.34EE252 pKa = 5.92GEE254 pKa = 4.25MNDD257 pKa = 3.53THH259 pKa = 6.3ITTAFISKK267 pKa = 10.05CSEE270 pKa = 3.67AAKK273 pKa = 10.22KK274 pKa = 10.14HH275 pKa = 4.83STTFSGLSARR285 pKa = 11.84VPAASGKK292 pKa = 8.93FDD294 pKa = 3.8EE295 pKa = 5.27VILVKK300 pKa = 10.55KK301 pKa = 9.48SAWGDD306 pKa = 3.49VPLFRR311 pKa = 11.84IDD313 pKa = 2.93EE314 pKa = 4.2ATRR317 pKa = 11.84KK318 pKa = 9.83FEE320 pKa = 4.01FMAIGTSTQVLCVKK334 pKa = 10.72DD335 pKa = 3.94DD336 pKa = 4.03LSYY339 pKa = 10.88MLLPTAHH346 pKa = 7.07
MM1 pKa = 7.29AQEE4 pKa = 3.93QVQVKK9 pKa = 10.12RR10 pKa = 11.84KK11 pKa = 6.29FTRR14 pKa = 11.84TICVLDD20 pKa = 4.26PGAKK24 pKa = 8.56TFCGKK29 pKa = 8.83VAKK32 pKa = 10.7LEE34 pKa = 4.23GKK36 pKa = 8.31TYY38 pKa = 10.32FVIKK42 pKa = 10.24IGRR45 pKa = 11.84TTQVGVSNTPVPKK58 pKa = 10.38CYY60 pKa = 10.56VLDD63 pKa = 3.72VAGPGSYY70 pKa = 9.06RR71 pKa = 11.84VQDD74 pKa = 3.8GQDD77 pKa = 3.18TVSVILSEE85 pKa = 4.49SGFEE89 pKa = 4.01ATGEE93 pKa = 3.84RR94 pKa = 11.84WEE96 pKa = 4.01EE97 pKa = 3.52WRR99 pKa = 11.84FEE101 pKa = 4.21VLSATPIVARR111 pKa = 11.84MAVGEE116 pKa = 4.59DD117 pKa = 3.48EE118 pKa = 4.68FDD120 pKa = 4.63AEE122 pKa = 4.27IKK124 pKa = 10.1YY125 pKa = 10.88SKK127 pKa = 10.81GLGSVPPYY135 pKa = 10.79AKK137 pKa = 10.62NEE139 pKa = 3.91ADD141 pKa = 3.6RR142 pKa = 11.84RR143 pKa = 11.84MMPTLPGVKK152 pKa = 9.91ALDD155 pKa = 3.33MPIRR159 pKa = 11.84DD160 pKa = 3.98FRR162 pKa = 11.84QHH164 pKa = 5.8ARR166 pKa = 11.84EE167 pKa = 3.95HH168 pKa = 6.32RR169 pKa = 11.84DD170 pKa = 3.21EE171 pKa = 4.82VRR173 pKa = 11.84AQLDD177 pKa = 3.37RR178 pKa = 11.84AARR181 pKa = 11.84RR182 pKa = 11.84LDD184 pKa = 3.68PSSSRR189 pKa = 11.84LLGASVGLSEE199 pKa = 5.38EE200 pKa = 4.09IPIEE204 pKa = 4.22SVLPQAKK211 pKa = 9.63RR212 pKa = 11.84EE213 pKa = 4.1PVVFASASRR222 pKa = 11.84EE223 pKa = 3.81LDD225 pKa = 2.96DD226 pKa = 4.87WGFDD230 pKa = 3.73MQPKK234 pKa = 9.62TSGDD238 pKa = 3.27WGAKK242 pKa = 7.51TPIPEE247 pKa = 4.15VEE249 pKa = 4.07SDD251 pKa = 3.34EE252 pKa = 5.92GEE254 pKa = 4.25MNDD257 pKa = 3.53THH259 pKa = 6.3ITTAFISKK267 pKa = 10.05CSEE270 pKa = 3.67AAKK273 pKa = 10.22KK274 pKa = 10.14HH275 pKa = 4.83STTFSGLSARR285 pKa = 11.84VPAASGKK292 pKa = 8.93FDD294 pKa = 3.8EE295 pKa = 5.27VILVKK300 pKa = 10.55KK301 pKa = 9.48SAWGDD306 pKa = 3.49VPLFRR311 pKa = 11.84IDD313 pKa = 2.93EE314 pKa = 4.2ATRR317 pKa = 11.84KK318 pKa = 9.83FEE320 pKa = 4.01FMAIGTSTQVLCVKK334 pKa = 10.72DD335 pKa = 3.94DD336 pKa = 4.03LSYY339 pKa = 10.88MLLPTAHH346 pKa = 7.07
Molecular weight: 38.19 kDa
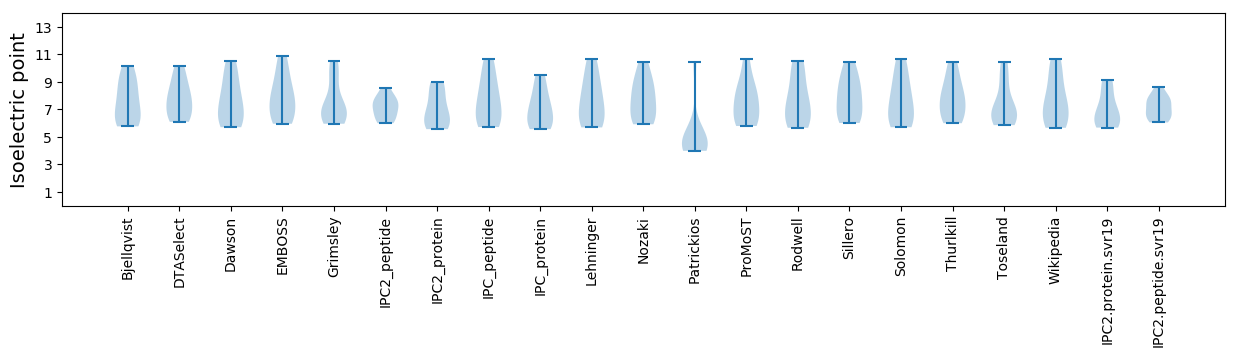
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5QLZ0|W5QLZ0_9REOV Outer capsid protein VP2 OS=Lebombo virus OX=40057 PE=3 SV=1
MM1 pKa = 7.79EE2 pKa = 5.12EE3 pKa = 4.32RR4 pKa = 11.84SQEE7 pKa = 4.03TPLTRR12 pKa = 11.84EE13 pKa = 3.77EE14 pKa = 4.24EE15 pKa = 4.32RR16 pKa = 11.84IKK18 pKa = 10.17MLRR21 pKa = 11.84RR22 pKa = 11.84EE23 pKa = 3.95VDD25 pKa = 2.98RR26 pKa = 11.84LLNLQIGRR34 pKa = 11.84NWEE37 pKa = 4.2AGTDD41 pKa = 3.28WMLAQIDD48 pKa = 3.96VLLFRR53 pKa = 11.84IRR55 pKa = 11.84VGNIKK60 pKa = 9.83QAEE63 pKa = 4.08EE64 pKa = 3.77QLLGMRR70 pKa = 11.84DD71 pKa = 3.53RR72 pKa = 11.84LEE74 pKa = 4.35DD75 pKa = 3.41ALEE78 pKa = 4.03RR79 pKa = 11.84KK80 pKa = 9.16RR81 pKa = 11.84RR82 pKa = 11.84GWRR85 pKa = 11.84VVWPPRR91 pKa = 11.84RR92 pKa = 3.51
MM1 pKa = 7.79EE2 pKa = 5.12EE3 pKa = 4.32RR4 pKa = 11.84SQEE7 pKa = 4.03TPLTRR12 pKa = 11.84EE13 pKa = 3.77EE14 pKa = 4.24EE15 pKa = 4.32RR16 pKa = 11.84IKK18 pKa = 10.17MLRR21 pKa = 11.84RR22 pKa = 11.84EE23 pKa = 3.95VDD25 pKa = 2.98RR26 pKa = 11.84LLNLQIGRR34 pKa = 11.84NWEE37 pKa = 4.2AGTDD41 pKa = 3.28WMLAQIDD48 pKa = 3.96VLLFRR53 pKa = 11.84IRR55 pKa = 11.84VGNIKK60 pKa = 9.83QAEE63 pKa = 4.08EE64 pKa = 3.77QLLGMRR70 pKa = 11.84DD71 pKa = 3.53RR72 pKa = 11.84LEE74 pKa = 4.35DD75 pKa = 3.41ALEE78 pKa = 4.03RR79 pKa = 11.84KK80 pKa = 9.16RR81 pKa = 11.84RR82 pKa = 11.84GWRR85 pKa = 11.84VVWPPRR91 pKa = 11.84RR92 pKa = 3.51
Molecular weight: 11.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6275 |
92 |
1298 |
570.5 |
64.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.299 ± 0.555 | 1.004 ± 0.268 |
6.167 ± 0.253 | 6.741 ± 0.401 |
3.952 ± 0.316 | 6.279 ± 0.411 |
2.327 ± 0.258 | 5.833 ± 0.275 |
5.546 ± 0.442 | 9.004 ± 0.316 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.474 ± 0.194 | 3.618 ± 0.271 |
4.335 ± 0.356 | 3.283 ± 0.344 |
7.363 ± 0.368 | 6.406 ± 0.449 |
5.243 ± 0.338 | 7.251 ± 0.184 |
1.386 ± 0.27 | 3.49 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
