
Erythrura gouldiae polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Gammapolyomavirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
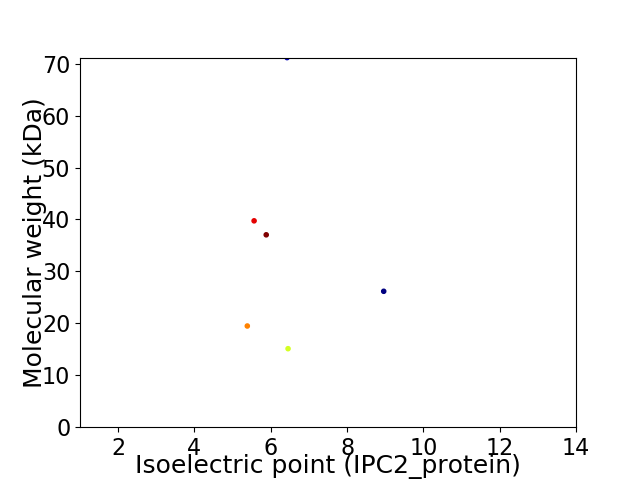
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1ETR3|A0A0K1ETR3_9POLY Large T antigen OS=Erythrura gouldiae polyomavirus 1 OX=2169903 PE=4 SV=1
MM1 pKa = 7.45SKK3 pKa = 11.07ADD5 pKa = 3.27IAEE8 pKa = 4.03LRR10 pKa = 11.84EE11 pKa = 4.0LLQLPKK17 pKa = 10.39NASFEE22 pKa = 4.48KK23 pKa = 10.43IKK25 pKa = 10.69AAYY28 pKa = 8.77RR29 pKa = 11.84KK30 pKa = 9.1KK31 pKa = 10.53ALMYY35 pKa = 10.6HH36 pKa = 6.72PDD38 pKa = 3.36KK39 pKa = 11.61GGDD42 pKa = 3.52EE43 pKa = 4.35EE44 pKa = 5.1KK45 pKa = 10.31MKK47 pKa = 10.69RR48 pKa = 11.84LNSLMEE54 pKa = 3.69QAKK57 pKa = 9.26TYY59 pKa = 10.91DD60 pKa = 3.72FSAEE64 pKa = 3.81NRR66 pKa = 11.84EE67 pKa = 3.63EE68 pKa = 4.46DD69 pKa = 4.22LYY71 pKa = 11.48CEE73 pKa = 4.42EE74 pKa = 4.52SLSSEE79 pKa = 4.32EE80 pKa = 4.46EE81 pKa = 4.17DD82 pKa = 3.77VSPGPSQRR90 pKa = 11.84AEE92 pKa = 3.82DD93 pKa = 3.31TGYY96 pKa = 11.49GSFSASFNASFTDD109 pKa = 3.66SVSALEE115 pKa = 4.22FQHH118 pKa = 6.95ACTKK122 pKa = 8.79ITKK125 pKa = 9.89LQDD128 pKa = 2.87KK129 pKa = 9.85LLAYY133 pKa = 9.32FAYY136 pKa = 10.78VDD138 pKa = 3.97GQRR141 pKa = 11.84KK142 pKa = 8.03QCLAQPYY149 pKa = 9.37RR150 pKa = 11.84DD151 pKa = 3.95LKK153 pKa = 10.58RR154 pKa = 11.84AYY156 pKa = 10.18QGVPWNLFNEE166 pKa = 4.7HH167 pKa = 5.37SHH169 pKa = 6.08FF170 pKa = 4.44
MM1 pKa = 7.45SKK3 pKa = 11.07ADD5 pKa = 3.27IAEE8 pKa = 4.03LRR10 pKa = 11.84EE11 pKa = 4.0LLQLPKK17 pKa = 10.39NASFEE22 pKa = 4.48KK23 pKa = 10.43IKK25 pKa = 10.69AAYY28 pKa = 8.77RR29 pKa = 11.84KK30 pKa = 9.1KK31 pKa = 10.53ALMYY35 pKa = 10.6HH36 pKa = 6.72PDD38 pKa = 3.36KK39 pKa = 11.61GGDD42 pKa = 3.52EE43 pKa = 4.35EE44 pKa = 5.1KK45 pKa = 10.31MKK47 pKa = 10.69RR48 pKa = 11.84LNSLMEE54 pKa = 3.69QAKK57 pKa = 9.26TYY59 pKa = 10.91DD60 pKa = 3.72FSAEE64 pKa = 3.81NRR66 pKa = 11.84EE67 pKa = 3.63EE68 pKa = 4.46DD69 pKa = 4.22LYY71 pKa = 11.48CEE73 pKa = 4.42EE74 pKa = 4.52SLSSEE79 pKa = 4.32EE80 pKa = 4.46EE81 pKa = 4.17DD82 pKa = 3.77VSPGPSQRR90 pKa = 11.84AEE92 pKa = 3.82DD93 pKa = 3.31TGYY96 pKa = 11.49GSFSASFNASFTDD109 pKa = 3.66SVSALEE115 pKa = 4.22FQHH118 pKa = 6.95ACTKK122 pKa = 8.79ITKK125 pKa = 9.89LQDD128 pKa = 2.87KK129 pKa = 9.85LLAYY133 pKa = 9.32FAYY136 pKa = 10.78VDD138 pKa = 3.97GQRR141 pKa = 11.84KK142 pKa = 8.03QCLAQPYY149 pKa = 9.37RR150 pKa = 11.84DD151 pKa = 3.95LKK153 pKa = 10.58RR154 pKa = 11.84AYY156 pKa = 10.18QGVPWNLFNEE166 pKa = 4.7HH167 pKa = 5.37SHH169 pKa = 6.08FF170 pKa = 4.44
Molecular weight: 19.49 kDa
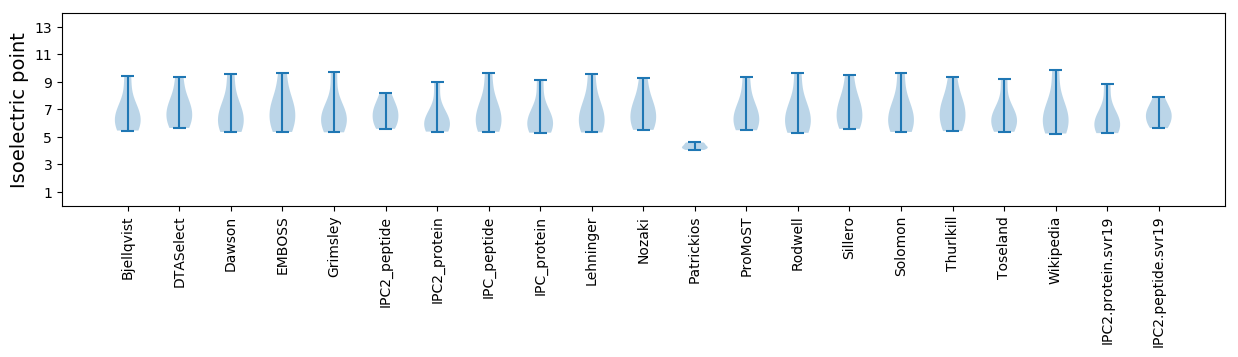
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1EUA9|A0A0K1EUA9_9POLY Capsid protein VP1 OS=Erythrura gouldiae polyomavirus 1 OX=2169903 PE=3 SV=1
MM1 pKa = 7.4AVAVWRR7 pKa = 11.84PEE9 pKa = 3.54VDD11 pKa = 4.04FLFPGASEE19 pKa = 4.8LVDD22 pKa = 3.25ALYY25 pKa = 10.79YY26 pKa = 10.27INPLEE31 pKa = 4.27WGPTIFQNIGRR42 pKa = 11.84YY43 pKa = 8.0IWDD46 pKa = 3.48YY47 pKa = 11.12VIQTGRR53 pKa = 11.84RR54 pKa = 11.84QLGEE58 pKa = 3.67ATRR61 pKa = 11.84AIVQTTSSNLYY72 pKa = 10.41DD73 pKa = 3.91LAARR77 pKa = 11.84AAEE80 pKa = 3.92RR81 pKa = 11.84ATWYY85 pKa = 8.45ITEE88 pKa = 4.18GPIRR92 pKa = 11.84SYY94 pKa = 11.33RR95 pKa = 11.84FLEE98 pKa = 4.47DD99 pKa = 3.8YY100 pKa = 11.36YY101 pKa = 10.85MEE103 pKa = 4.85LPILRR108 pKa = 11.84PDD110 pKa = 3.28ARR112 pKa = 11.84RR113 pKa = 11.84GRR115 pKa = 11.84ALAIGSDD122 pKa = 3.8AEE124 pKa = 4.28EE125 pKa = 4.48EE126 pKa = 4.1EE127 pKa = 4.59LPEE130 pKa = 4.46GLRR133 pKa = 11.84KK134 pKa = 9.72LHH136 pKa = 6.64PKK138 pKa = 7.77QTHH141 pKa = 5.77EE142 pKa = 4.07PTGATVAPSVAPGGANQRR160 pKa = 11.84HH161 pKa = 6.0CPDD164 pKa = 2.74WMLPLILGLYY174 pKa = 8.72GVVYY178 pKa = 9.34PGWKK182 pKa = 10.28AEE184 pKa = 3.97VEE186 pKa = 3.99LLEE189 pKa = 5.21KK190 pKa = 10.88EE191 pKa = 4.44EE192 pKa = 4.22SQHH195 pKa = 5.9GSKK198 pKa = 9.2TGPYY202 pKa = 8.8RR203 pKa = 11.84RR204 pKa = 11.84SRR206 pKa = 11.84SKK208 pKa = 10.55PRR210 pKa = 11.84SKK212 pKa = 9.45TPYY215 pKa = 8.45QRR217 pKa = 11.84RR218 pKa = 11.84HH219 pKa = 5.39RR220 pKa = 11.84SSRR223 pKa = 11.84SKK225 pKa = 10.43VRR227 pKa = 11.84SRR229 pKa = 3.57
MM1 pKa = 7.4AVAVWRR7 pKa = 11.84PEE9 pKa = 3.54VDD11 pKa = 4.04FLFPGASEE19 pKa = 4.8LVDD22 pKa = 3.25ALYY25 pKa = 10.79YY26 pKa = 10.27INPLEE31 pKa = 4.27WGPTIFQNIGRR42 pKa = 11.84YY43 pKa = 8.0IWDD46 pKa = 3.48YY47 pKa = 11.12VIQTGRR53 pKa = 11.84RR54 pKa = 11.84QLGEE58 pKa = 3.67ATRR61 pKa = 11.84AIVQTTSSNLYY72 pKa = 10.41DD73 pKa = 3.91LAARR77 pKa = 11.84AAEE80 pKa = 3.92RR81 pKa = 11.84ATWYY85 pKa = 8.45ITEE88 pKa = 4.18GPIRR92 pKa = 11.84SYY94 pKa = 11.33RR95 pKa = 11.84FLEE98 pKa = 4.47DD99 pKa = 3.8YY100 pKa = 11.36YY101 pKa = 10.85MEE103 pKa = 4.85LPILRR108 pKa = 11.84PDD110 pKa = 3.28ARR112 pKa = 11.84RR113 pKa = 11.84GRR115 pKa = 11.84ALAIGSDD122 pKa = 3.8AEE124 pKa = 4.28EE125 pKa = 4.48EE126 pKa = 4.1EE127 pKa = 4.59LPEE130 pKa = 4.46GLRR133 pKa = 11.84KK134 pKa = 9.72LHH136 pKa = 6.64PKK138 pKa = 7.77QTHH141 pKa = 5.77EE142 pKa = 4.07PTGATVAPSVAPGGANQRR160 pKa = 11.84HH161 pKa = 6.0CPDD164 pKa = 2.74WMLPLILGLYY174 pKa = 8.72GVVYY178 pKa = 9.34PGWKK182 pKa = 10.28AEE184 pKa = 3.97VEE186 pKa = 3.99LLEE189 pKa = 5.21KK190 pKa = 10.88EE191 pKa = 4.44EE192 pKa = 4.22SQHH195 pKa = 5.9GSKK198 pKa = 9.2TGPYY202 pKa = 8.8RR203 pKa = 11.84RR204 pKa = 11.84SRR206 pKa = 11.84SKK208 pKa = 10.55PRR210 pKa = 11.84SKK212 pKa = 9.45TPYY215 pKa = 8.45QRR217 pKa = 11.84RR218 pKa = 11.84HH219 pKa = 5.39RR220 pKa = 11.84SSRR223 pKa = 11.84SKK225 pKa = 10.43VRR227 pKa = 11.84SRR229 pKa = 3.57
Molecular weight: 26.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1878 |
144 |
634 |
313.0 |
34.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.253 ± 0.927 | 1.651 ± 0.497 |
4.739 ± 0.491 | 7.295 ± 0.533 |
3.248 ± 0.443 | 7.401 ± 0.781 |
2.13 ± 0.27 | 4.313 ± 0.542 |
5.325 ± 0.872 | 10.064 ± 0.444 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.917 ± 0.265 | 3.994 ± 0.6 |
7.295 ± 1.142 | 3.834 ± 0.179 |
6.337 ± 0.762 | 6.55 ± 0.53 |
6.07 ± 0.359 | 4.846 ± 0.638 |
1.225 ± 0.326 | 3.514 ± 0.497 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
