
Bole Tick Virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Alpharicinrhavirus; Bole alpharicinrhavirus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
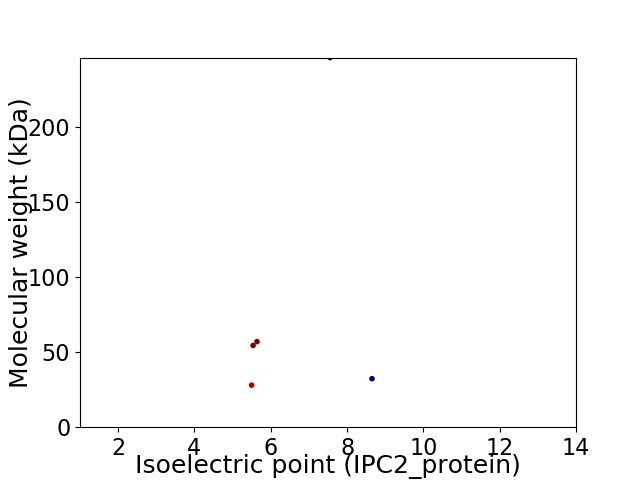
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
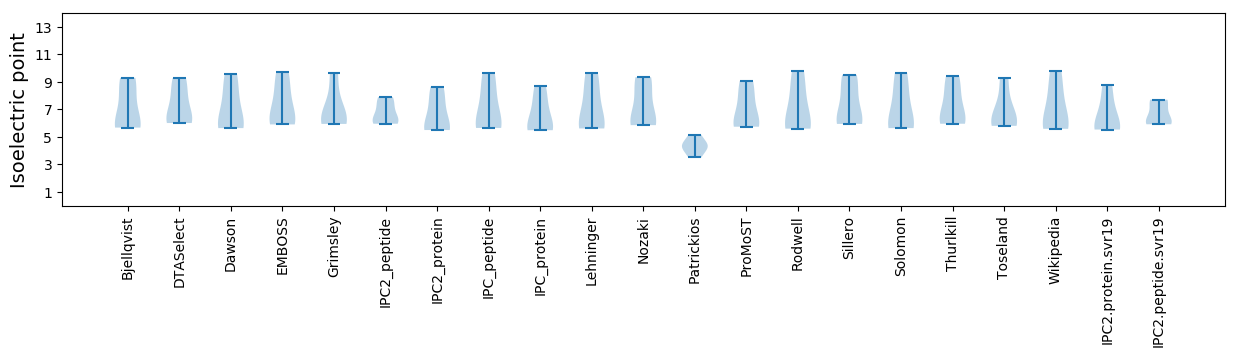
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KTD4|A0A0B5KTD4_9RHAB GDP polyribonucleotidyltransferase OS=Bole Tick Virus 2 OX=1608041 GN=L PE=4 SV=1
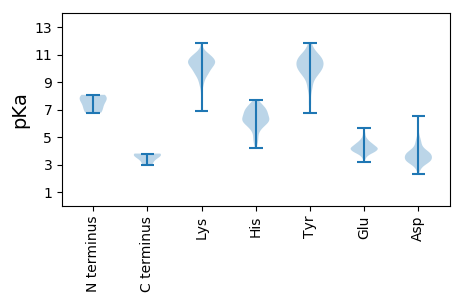
MM1 pKa = 6.77GTVGEE6 pKa = 4.39VFLLLSLLSFLPLAGSDD23 pKa = 3.65PVVKK27 pKa = 10.56AIAAPYY33 pKa = 8.57FFPEE37 pKa = 3.82NLNYY41 pKa = 10.0AWHH44 pKa = 6.99PIEE47 pKa = 4.53VTSLTCPPQRR57 pKa = 11.84SIPEE61 pKa = 3.84QDD63 pKa = 2.92NGIPVIFEE71 pKa = 4.44TIHH74 pKa = 6.0PSSLEE79 pKa = 3.74RR80 pKa = 11.84ALVNGYY86 pKa = 9.5SCYY89 pKa = 9.8TSTMAVKK96 pKa = 10.5CSVNFVGWKK105 pKa = 7.95TLSHH109 pKa = 6.44QITNKK114 pKa = 9.68EE115 pKa = 4.06PSSTTCWEE123 pKa = 4.28AIKK126 pKa = 10.37RR127 pKa = 11.84QEE129 pKa = 5.28DD130 pKa = 4.44GAQSPPPTFPAPNCAWWSEE149 pKa = 3.65NWAEE153 pKa = 4.46LDD155 pKa = 3.44YY156 pKa = 11.35TLVLKK161 pKa = 10.83HH162 pKa = 6.54PARR165 pKa = 11.84QDD167 pKa = 3.26PYY169 pKa = 11.13TEE171 pKa = 4.22ALYY174 pKa = 10.88DD175 pKa = 3.69PLFPGGSCNKK185 pKa = 9.87AEE187 pKa = 4.98CPLIHH192 pKa = 7.71DD193 pKa = 4.44GGIWIQTEE201 pKa = 4.2PLASICKK208 pKa = 9.35HH209 pKa = 4.86WEE211 pKa = 3.72VLQGLTYY218 pKa = 9.75TGPEE222 pKa = 3.32IGRR225 pKa = 11.84ILFSPEE231 pKa = 3.93GPPKK235 pKa = 10.7YY236 pKa = 9.76LDD238 pKa = 3.31HH239 pKa = 6.71SCRR242 pKa = 11.84MTFCGRR248 pKa = 11.84RR249 pKa = 11.84GYY251 pKa = 10.6RR252 pKa = 11.84LQDD255 pKa = 3.23GEE257 pKa = 4.59FLVFSSPPAWGVPPVCPAGTLVRR280 pKa = 11.84AHH282 pKa = 5.98TPEE285 pKa = 4.31EE286 pKa = 3.96EE287 pKa = 4.18IRR289 pKa = 11.84WNEE292 pKa = 3.45ISKK295 pKa = 9.48MEE297 pKa = 4.06EE298 pKa = 3.9ADD300 pKa = 3.97RR301 pKa = 11.84LMCISRR307 pKa = 11.84LSVAYY312 pKa = 8.41ATGKK316 pKa = 10.52VSLEE320 pKa = 3.95LLGSLVPSHH329 pKa = 6.75GGPGTAYY336 pKa = 10.41RR337 pKa = 11.84INNGTLEE344 pKa = 4.32AAHH347 pKa = 6.05VKK349 pKa = 9.47YY350 pKa = 10.88VPLINSSNEE359 pKa = 3.57EE360 pKa = 3.52GDD362 pKa = 4.28LIGVGPDD369 pKa = 3.2GTPILWEE376 pKa = 3.88YY377 pKa = 9.65WVLSGSRR384 pKa = 11.84IIGPNGVYY392 pKa = 9.96KK393 pKa = 10.65SKK395 pKa = 10.81GRR397 pKa = 11.84IIVPNFEE404 pKa = 4.24RR405 pKa = 11.84RR406 pKa = 11.84KK407 pKa = 8.79LTYY410 pKa = 10.5DD411 pKa = 3.3LTIHH415 pKa = 5.87VFEE418 pKa = 5.08DD419 pKa = 3.72LKK421 pKa = 10.98EE422 pKa = 4.22IPHH425 pKa = 6.9PSLVIRR431 pKa = 11.84SNHH434 pKa = 5.09TDD436 pKa = 3.37LLRR439 pKa = 11.84KK440 pKa = 9.35VSHH443 pKa = 5.67NQGVEE448 pKa = 3.77GDD450 pKa = 3.26HH451 pKa = 6.35WASIRR456 pKa = 11.84LWFSSLWGSFIWTCGLALIGLILICCICRR485 pKa = 11.84RR486 pKa = 11.84VRR488 pKa = 11.84CCCRR492 pKa = 11.84GCGRR496 pKa = 11.84PQSKK500 pKa = 9.59EE501 pKa = 3.59AGGWEE506 pKa = 4.29SIEE509 pKa = 4.13MNDD512 pKa = 4.26LL513 pKa = 3.69
MM1 pKa = 6.77GTVGEE6 pKa = 4.39VFLLLSLLSFLPLAGSDD23 pKa = 3.65PVVKK27 pKa = 10.56AIAAPYY33 pKa = 8.57FFPEE37 pKa = 3.82NLNYY41 pKa = 10.0AWHH44 pKa = 6.99PIEE47 pKa = 4.53VTSLTCPPQRR57 pKa = 11.84SIPEE61 pKa = 3.84QDD63 pKa = 2.92NGIPVIFEE71 pKa = 4.44TIHH74 pKa = 6.0PSSLEE79 pKa = 3.74RR80 pKa = 11.84ALVNGYY86 pKa = 9.5SCYY89 pKa = 9.8TSTMAVKK96 pKa = 10.5CSVNFVGWKK105 pKa = 7.95TLSHH109 pKa = 6.44QITNKK114 pKa = 9.68EE115 pKa = 4.06PSSTTCWEE123 pKa = 4.28AIKK126 pKa = 10.37RR127 pKa = 11.84QEE129 pKa = 5.28DD130 pKa = 4.44GAQSPPPTFPAPNCAWWSEE149 pKa = 3.65NWAEE153 pKa = 4.46LDD155 pKa = 3.44YY156 pKa = 11.35TLVLKK161 pKa = 10.83HH162 pKa = 6.54PARR165 pKa = 11.84QDD167 pKa = 3.26PYY169 pKa = 11.13TEE171 pKa = 4.22ALYY174 pKa = 10.88DD175 pKa = 3.69PLFPGGSCNKK185 pKa = 9.87AEE187 pKa = 4.98CPLIHH192 pKa = 7.71DD193 pKa = 4.44GGIWIQTEE201 pKa = 4.2PLASICKK208 pKa = 9.35HH209 pKa = 4.86WEE211 pKa = 3.72VLQGLTYY218 pKa = 9.75TGPEE222 pKa = 3.32IGRR225 pKa = 11.84ILFSPEE231 pKa = 3.93GPPKK235 pKa = 10.7YY236 pKa = 9.76LDD238 pKa = 3.31HH239 pKa = 6.71SCRR242 pKa = 11.84MTFCGRR248 pKa = 11.84RR249 pKa = 11.84GYY251 pKa = 10.6RR252 pKa = 11.84LQDD255 pKa = 3.23GEE257 pKa = 4.59FLVFSSPPAWGVPPVCPAGTLVRR280 pKa = 11.84AHH282 pKa = 5.98TPEE285 pKa = 4.31EE286 pKa = 3.96EE287 pKa = 4.18IRR289 pKa = 11.84WNEE292 pKa = 3.45ISKK295 pKa = 9.48MEE297 pKa = 4.06EE298 pKa = 3.9ADD300 pKa = 3.97RR301 pKa = 11.84LMCISRR307 pKa = 11.84LSVAYY312 pKa = 8.41ATGKK316 pKa = 10.52VSLEE320 pKa = 3.95LLGSLVPSHH329 pKa = 6.75GGPGTAYY336 pKa = 10.41RR337 pKa = 11.84INNGTLEE344 pKa = 4.32AAHH347 pKa = 6.05VKK349 pKa = 9.47YY350 pKa = 10.88VPLINSSNEE359 pKa = 3.57EE360 pKa = 3.52GDD362 pKa = 4.28LIGVGPDD369 pKa = 3.2GTPILWEE376 pKa = 3.88YY377 pKa = 9.65WVLSGSRR384 pKa = 11.84IIGPNGVYY392 pKa = 9.96KK393 pKa = 10.65SKK395 pKa = 10.81GRR397 pKa = 11.84IIVPNFEE404 pKa = 4.24RR405 pKa = 11.84RR406 pKa = 11.84KK407 pKa = 8.79LTYY410 pKa = 10.5DD411 pKa = 3.3LTIHH415 pKa = 5.87VFEE418 pKa = 5.08DD419 pKa = 3.72LKK421 pKa = 10.98EE422 pKa = 4.22IPHH425 pKa = 6.9PSLVIRR431 pKa = 11.84SNHH434 pKa = 5.09TDD436 pKa = 3.37LLRR439 pKa = 11.84KK440 pKa = 9.35VSHH443 pKa = 5.67NQGVEE448 pKa = 3.77GDD450 pKa = 3.26HH451 pKa = 6.35WASIRR456 pKa = 11.84LWFSSLWGSFIWTCGLALIGLILICCICRR485 pKa = 11.84RR486 pKa = 11.84VRR488 pKa = 11.84CCCRR492 pKa = 11.84GCGRR496 pKa = 11.84PQSKK500 pKa = 9.59EE501 pKa = 3.59AGGWEE506 pKa = 4.29SIEE509 pKa = 4.13MNDD512 pKa = 4.26LL513 pKa = 3.69
Molecular weight: 57.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KRE4|A0A0B5KRE4_9RHAB Glycoprotein OS=Bole Tick Virus 2 OX=1608041 GN=G PE=4 SV=1
MM1 pKa = 7.12QYY3 pKa = 10.53SWTKK7 pKa = 11.18DD8 pKa = 3.22LMFLFSNPLIFIFRR22 pKa = 11.84PILHH26 pKa = 7.0EE27 pKa = 4.47KK28 pKa = 8.75NQQISITMWRR38 pKa = 11.84FWRR41 pKa = 11.84VKK43 pKa = 10.24SPQPPPIPVEE53 pKa = 4.06EE54 pKa = 4.43EE55 pKa = 3.89EE56 pKa = 4.47EE57 pKa = 4.03EE58 pKa = 4.54DD59 pKa = 4.64LFGQIPSAPRR69 pKa = 11.84PKK71 pKa = 10.49GLRR74 pKa = 11.84ALLQGWRR81 pKa = 11.84ARR83 pKa = 11.84TRR85 pKa = 11.84STRR88 pKa = 11.84PSVHH92 pKa = 6.35ARR94 pKa = 11.84PDD96 pKa = 3.37LWAHH100 pKa = 6.66PLDD103 pKa = 4.96DD104 pKa = 6.17LPNTWTGLAVPNLPSEE120 pKa = 4.41KK121 pKa = 9.42PEE123 pKa = 4.23HH124 pKa = 5.45VTLDD128 pKa = 3.05IAATLEE134 pKa = 4.0VRR136 pKa = 11.84APRR139 pKa = 11.84PLDD142 pKa = 3.34QKK144 pKa = 9.67TLAEE148 pKa = 4.07MSLNFPLAYY157 pKa = 9.95QGEE160 pKa = 4.53ARR162 pKa = 11.84SKK164 pKa = 10.41PLFSVLLAHH173 pKa = 6.92LVSKK177 pKa = 11.02ASAHH181 pKa = 4.18QTRR184 pKa = 11.84SGLIYY189 pKa = 10.32SASLTEE195 pKa = 4.4CVIFTGRR202 pKa = 11.84KK203 pKa = 8.86GFRR206 pKa = 11.84PDD208 pKa = 3.73NRR210 pKa = 11.84LQTYY214 pKa = 9.54QDD216 pKa = 3.42TYY218 pKa = 7.21EE219 pKa = 3.75WRR221 pKa = 11.84YY222 pKa = 9.48KK223 pKa = 10.38GQVFFWKK230 pKa = 10.27LRR232 pKa = 11.84FEE234 pKa = 4.22AHH236 pKa = 5.43PTVMKK241 pKa = 10.18GIPLHH246 pKa = 6.14QILSVEE252 pKa = 4.1VMPLLRR258 pKa = 11.84ALGLHH263 pKa = 4.62PHH265 pKa = 5.67FRR267 pKa = 11.84KK268 pKa = 10.22EE269 pKa = 3.85EE270 pKa = 3.91QYY272 pKa = 11.53LEE274 pKa = 4.12IEE276 pKa = 4.92GPQQ279 pKa = 2.99
MM1 pKa = 7.12QYY3 pKa = 10.53SWTKK7 pKa = 11.18DD8 pKa = 3.22LMFLFSNPLIFIFRR22 pKa = 11.84PILHH26 pKa = 7.0EE27 pKa = 4.47KK28 pKa = 8.75NQQISITMWRR38 pKa = 11.84FWRR41 pKa = 11.84VKK43 pKa = 10.24SPQPPPIPVEE53 pKa = 4.06EE54 pKa = 4.43EE55 pKa = 3.89EE56 pKa = 4.47EE57 pKa = 4.03EE58 pKa = 4.54DD59 pKa = 4.64LFGQIPSAPRR69 pKa = 11.84PKK71 pKa = 10.49GLRR74 pKa = 11.84ALLQGWRR81 pKa = 11.84ARR83 pKa = 11.84TRR85 pKa = 11.84STRR88 pKa = 11.84PSVHH92 pKa = 6.35ARR94 pKa = 11.84PDD96 pKa = 3.37LWAHH100 pKa = 6.66PLDD103 pKa = 4.96DD104 pKa = 6.17LPNTWTGLAVPNLPSEE120 pKa = 4.41KK121 pKa = 9.42PEE123 pKa = 4.23HH124 pKa = 5.45VTLDD128 pKa = 3.05IAATLEE134 pKa = 4.0VRR136 pKa = 11.84APRR139 pKa = 11.84PLDD142 pKa = 3.34QKK144 pKa = 9.67TLAEE148 pKa = 4.07MSLNFPLAYY157 pKa = 9.95QGEE160 pKa = 4.53ARR162 pKa = 11.84SKK164 pKa = 10.41PLFSVLLAHH173 pKa = 6.92LVSKK177 pKa = 11.02ASAHH181 pKa = 4.18QTRR184 pKa = 11.84SGLIYY189 pKa = 10.32SASLTEE195 pKa = 4.4CVIFTGRR202 pKa = 11.84KK203 pKa = 8.86GFRR206 pKa = 11.84PDD208 pKa = 3.73NRR210 pKa = 11.84LQTYY214 pKa = 9.54QDD216 pKa = 3.42TYY218 pKa = 7.21EE219 pKa = 3.75WRR221 pKa = 11.84YY222 pKa = 9.48KK223 pKa = 10.38GQVFFWKK230 pKa = 10.27LRR232 pKa = 11.84FEE234 pKa = 4.22AHH236 pKa = 5.43PTVMKK241 pKa = 10.18GIPLHH246 pKa = 6.14QILSVEE252 pKa = 4.1VMPLLRR258 pKa = 11.84ALGLHH263 pKa = 4.62PHH265 pKa = 5.67FRR267 pKa = 11.84KK268 pKa = 10.22EE269 pKa = 3.85EE270 pKa = 3.91QYY272 pKa = 11.53LEE274 pKa = 4.12IEE276 pKa = 4.92GPQQ279 pKa = 2.99
Molecular weight: 32.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3703 |
255 |
2171 |
740.6 |
83.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.076 ± 0.707 | 1.512 ± 0.56 |
4.807 ± 0.562 | 6.427 ± 0.366 |
4.537 ± 0.578 | 6.562 ± 0.569 |
2.565 ± 0.207 | 5.752 ± 0.353 |
4.861 ± 0.42 | 10.883 ± 0.619 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.284 | 2.863 ± 0.303 |
6.67 ± 0.835 | 3.646 ± 0.391 |
6.184 ± 0.368 | 7.426 ± 0.686 |
5.806 ± 0.823 | 6.103 ± 0.454 |
2.079 ± 0.363 | 3.052 ± 0.168 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
