
Cellulophaga phage phi19:3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae; crAss-like viruses
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
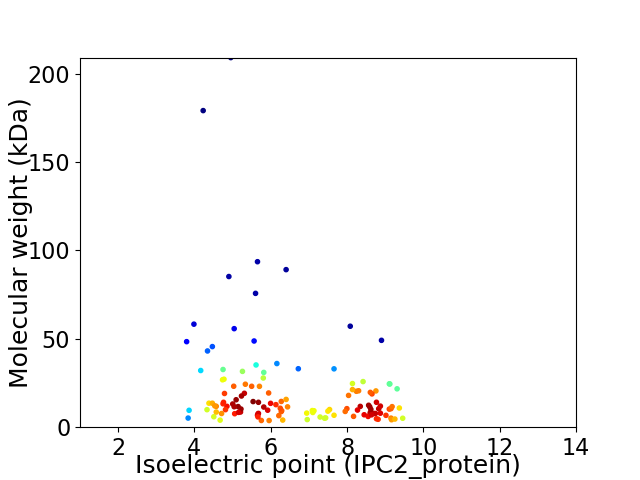
Virtual 2D-PAGE plot for 129 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9ZWL2|R9ZWL2_9CAUD Uncharacterized protein OS=Cellulophaga phage phi19:3 OX=1327971 GN=Phi19:3_gp092 PE=4 SV=1
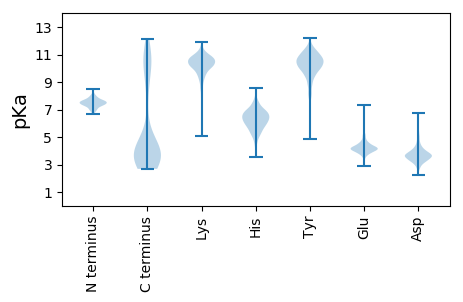
MM1 pKa = 7.47KK2 pKa = 10.61NKK4 pKa = 8.56ITIIMILMSISFGYY18 pKa = 9.85SQVLVPEE25 pKa = 4.2NTGYY29 pKa = 11.18GYY31 pKa = 11.06DD32 pKa = 3.4NLTTVEE38 pKa = 4.61LNALTKK44 pKa = 10.63KK45 pKa = 10.5KK46 pKa = 10.94VGDD49 pKa = 3.23IYY51 pKa = 11.59YY52 pKa = 10.48NADD55 pKa = 3.03LGYY58 pKa = 9.03HH59 pKa = 5.2VKK61 pKa = 9.49WNGVIFEE68 pKa = 4.6SLASGISGIDD78 pKa = 3.43PADD81 pKa = 3.37QDD83 pKa = 3.85KK84 pKa = 11.33LNNISITQPVNLDD97 pKa = 3.69TVEE100 pKa = 4.31SLANSALQTEE110 pKa = 4.96TDD112 pKa = 3.69PTTTPANIKK121 pKa = 10.45AKK123 pKa = 10.55LEE125 pKa = 4.19SLTGTNRR132 pKa = 11.84LDD134 pKa = 3.27AFAIKK139 pKa = 10.27NLSAQEE145 pKa = 3.96TDD147 pKa = 3.46PNLTKK152 pKa = 11.0SNVEE156 pKa = 3.97GLGISYY162 pKa = 9.46TSLSDD167 pKa = 3.3IPAVSSPLTFGPGFTRR183 pKa = 11.84TGDD186 pKa = 3.71NIVLDD191 pKa = 4.09NPFTTANEE199 pKa = 4.21TTLNSALQSEE209 pKa = 4.65TDD211 pKa = 3.7PLYY214 pKa = 10.96SAWDD218 pKa = 3.41KK219 pKa = 11.41DD220 pKa = 4.07YY221 pKa = 11.58NDD223 pKa = 5.36LINRR227 pKa = 11.84PVNLVTYY234 pKa = 10.27DD235 pKa = 3.75PLTAQDD241 pKa = 3.7VEE243 pKa = 4.39ISVLGGSDD251 pKa = 3.46SEE253 pKa = 4.81SYY255 pKa = 10.7ILEE258 pKa = 4.31FDD260 pKa = 3.96PNGYY264 pKa = 10.14DD265 pKa = 3.5YY266 pKa = 11.39SGEE269 pKa = 4.11SSGTGTVDD277 pKa = 2.9QTIIDD282 pKa = 4.09GSTNAVSGNAVFDD295 pKa = 4.11GLSSKK300 pKa = 11.05LNLSGGTMTGEE311 pKa = 4.38LDD313 pKa = 4.21LSLLMPSSVVSFIGSNTDD331 pKa = 3.35PFSSGSFKK339 pKa = 11.04DD340 pKa = 4.13FNVSDD345 pKa = 4.1GSTLIDD351 pKa = 3.56YY352 pKa = 8.76QFSPTSTALSIRR364 pKa = 11.84TNTLVNQTAYY374 pKa = 7.58TAKK377 pKa = 9.42YY378 pKa = 7.38TLPHH382 pKa = 6.52TGTPVNDD389 pKa = 3.52TDD391 pKa = 6.46LITKK395 pKa = 9.38VYY397 pKa = 11.2ADD399 pKa = 3.58ATYY402 pKa = 10.1GASGSYY408 pKa = 10.67EE409 pKa = 3.92EE410 pKa = 5.03GTFTLTNQDD419 pKa = 2.46TDD421 pKa = 3.69YY422 pKa = 11.9SFISQNNTFVRR433 pKa = 11.84FGDD436 pKa = 3.6LVYY439 pKa = 10.39YY440 pKa = 10.16KK441 pKa = 10.81VYY443 pKa = 9.09ITLNKK448 pKa = 7.14TTNTSDD454 pKa = 3.34VFFTLSSASTLPAHH468 pKa = 6.91ATQSVNNINPYY479 pKa = 10.26FSDD482 pKa = 3.94LLSTSGNLNANATALYY498 pKa = 8.82FQHH501 pKa = 6.67NGNVWYY507 pKa = 10.39LKK509 pKa = 10.28QSYY512 pKa = 9.93AGNIGVNVSNIDD524 pKa = 3.53STSSGNFVISFSGTYY539 pKa = 8.85LTDD542 pKa = 3.05
MM1 pKa = 7.47KK2 pKa = 10.61NKK4 pKa = 8.56ITIIMILMSISFGYY18 pKa = 9.85SQVLVPEE25 pKa = 4.2NTGYY29 pKa = 11.18GYY31 pKa = 11.06DD32 pKa = 3.4NLTTVEE38 pKa = 4.61LNALTKK44 pKa = 10.63KK45 pKa = 10.5KK46 pKa = 10.94VGDD49 pKa = 3.23IYY51 pKa = 11.59YY52 pKa = 10.48NADD55 pKa = 3.03LGYY58 pKa = 9.03HH59 pKa = 5.2VKK61 pKa = 9.49WNGVIFEE68 pKa = 4.6SLASGISGIDD78 pKa = 3.43PADD81 pKa = 3.37QDD83 pKa = 3.85KK84 pKa = 11.33LNNISITQPVNLDD97 pKa = 3.69TVEE100 pKa = 4.31SLANSALQTEE110 pKa = 4.96TDD112 pKa = 3.69PTTTPANIKK121 pKa = 10.45AKK123 pKa = 10.55LEE125 pKa = 4.19SLTGTNRR132 pKa = 11.84LDD134 pKa = 3.27AFAIKK139 pKa = 10.27NLSAQEE145 pKa = 3.96TDD147 pKa = 3.46PNLTKK152 pKa = 11.0SNVEE156 pKa = 3.97GLGISYY162 pKa = 9.46TSLSDD167 pKa = 3.3IPAVSSPLTFGPGFTRR183 pKa = 11.84TGDD186 pKa = 3.71NIVLDD191 pKa = 4.09NPFTTANEE199 pKa = 4.21TTLNSALQSEE209 pKa = 4.65TDD211 pKa = 3.7PLYY214 pKa = 10.96SAWDD218 pKa = 3.41KK219 pKa = 11.41DD220 pKa = 4.07YY221 pKa = 11.58NDD223 pKa = 5.36LINRR227 pKa = 11.84PVNLVTYY234 pKa = 10.27DD235 pKa = 3.75PLTAQDD241 pKa = 3.7VEE243 pKa = 4.39ISVLGGSDD251 pKa = 3.46SEE253 pKa = 4.81SYY255 pKa = 10.7ILEE258 pKa = 4.31FDD260 pKa = 3.96PNGYY264 pKa = 10.14DD265 pKa = 3.5YY266 pKa = 11.39SGEE269 pKa = 4.11SSGTGTVDD277 pKa = 2.9QTIIDD282 pKa = 4.09GSTNAVSGNAVFDD295 pKa = 4.11GLSSKK300 pKa = 11.05LNLSGGTMTGEE311 pKa = 4.38LDD313 pKa = 4.21LSLLMPSSVVSFIGSNTDD331 pKa = 3.35PFSSGSFKK339 pKa = 11.04DD340 pKa = 4.13FNVSDD345 pKa = 4.1GSTLIDD351 pKa = 3.56YY352 pKa = 8.76QFSPTSTALSIRR364 pKa = 11.84TNTLVNQTAYY374 pKa = 7.58TAKK377 pKa = 9.42YY378 pKa = 7.38TLPHH382 pKa = 6.52TGTPVNDD389 pKa = 3.52TDD391 pKa = 6.46LITKK395 pKa = 9.38VYY397 pKa = 11.2ADD399 pKa = 3.58ATYY402 pKa = 10.1GASGSYY408 pKa = 10.67EE409 pKa = 3.92EE410 pKa = 5.03GTFTLTNQDD419 pKa = 2.46TDD421 pKa = 3.69YY422 pKa = 11.9SFISQNNTFVRR433 pKa = 11.84FGDD436 pKa = 3.6LVYY439 pKa = 10.39YY440 pKa = 10.16KK441 pKa = 10.81VYY443 pKa = 9.09ITLNKK448 pKa = 7.14TTNTSDD454 pKa = 3.34VFFTLSSASTLPAHH468 pKa = 6.91ATQSVNNINPYY479 pKa = 10.26FSDD482 pKa = 3.94LLSTSGNLNANATALYY498 pKa = 8.82FQHH501 pKa = 6.67NGNVWYY507 pKa = 10.39LKK509 pKa = 10.28QSYY512 pKa = 9.93AGNIGVNVSNIDD524 pKa = 3.53STSSGNFVISFSGTYY539 pKa = 8.85LTDD542 pKa = 3.05
Molecular weight: 58.27 kDa
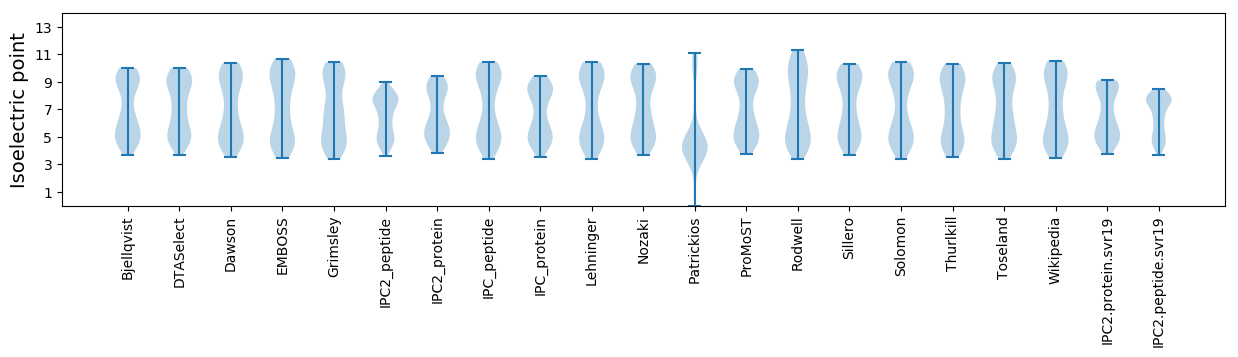
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9ZY34|R9ZY34_9CAUD Site specific recombinase tyrosine OS=Cellulophaga phage phi19:3 OX=1327971 GN=Phi19:3_gp036 PE=3 SV=1
MM1 pKa = 7.58EE2 pKa = 4.14IWKK5 pKa = 9.34PIKK8 pKa = 10.3GYY10 pKa = 10.09EE11 pKa = 4.1NIYY14 pKa = 9.89QVSNFGRR21 pKa = 11.84VKK23 pKa = 10.44SLDD26 pKa = 3.19RR27 pKa = 11.84IVFNKK32 pKa = 10.51GNGTRR37 pKa = 11.84CKK39 pKa = 10.16TKK41 pKa = 10.52GRR43 pKa = 11.84VLKK46 pKa = 10.45QSKK49 pKa = 10.59DD50 pKa = 3.22KK51 pKa = 11.39GGYY54 pKa = 10.14LYY56 pKa = 11.1VGLYY60 pKa = 10.7NKK62 pKa = 10.43DD63 pKa = 3.48NEE65 pKa = 4.48KK66 pKa = 9.6TSSIKK71 pKa = 8.59VHH73 pKa = 6.31RR74 pKa = 11.84LVAFSFCSGYY84 pKa = 9.2TEE86 pKa = 4.04GLEE89 pKa = 4.25VNHH92 pKa = 7.29KK93 pKa = 10.87DD94 pKa = 4.39GIRR97 pKa = 11.84DD98 pKa = 3.5NNLYY102 pKa = 10.26TNLEE106 pKa = 4.14WVTRR110 pKa = 11.84SQNIRR115 pKa = 11.84DD116 pKa = 3.62TYY118 pKa = 9.56KK119 pKa = 10.43RR120 pKa = 11.84GRR122 pKa = 11.84ITYY125 pKa = 7.32GQKK128 pKa = 10.74NNASKK133 pKa = 11.07LMDD136 pKa = 4.12RR137 pKa = 11.84DD138 pKa = 3.12IGVISSLYY146 pKa = 10.48DD147 pKa = 3.19SGVSQSIISLAFGVSQSTISNVIKK171 pKa = 10.62NKK173 pKa = 9.74HH174 pKa = 5.46YY175 pKa = 11.08KK176 pKa = 10.11NGLIEE181 pKa = 5.52KK182 pKa = 10.11GLADD186 pKa = 3.63QQRR189 pKa = 11.84TNN191 pKa = 3.23
MM1 pKa = 7.58EE2 pKa = 4.14IWKK5 pKa = 9.34PIKK8 pKa = 10.3GYY10 pKa = 10.09EE11 pKa = 4.1NIYY14 pKa = 9.89QVSNFGRR21 pKa = 11.84VKK23 pKa = 10.44SLDD26 pKa = 3.19RR27 pKa = 11.84IVFNKK32 pKa = 10.51GNGTRR37 pKa = 11.84CKK39 pKa = 10.16TKK41 pKa = 10.52GRR43 pKa = 11.84VLKK46 pKa = 10.45QSKK49 pKa = 10.59DD50 pKa = 3.22KK51 pKa = 11.39GGYY54 pKa = 10.14LYY56 pKa = 11.1VGLYY60 pKa = 10.7NKK62 pKa = 10.43DD63 pKa = 3.48NEE65 pKa = 4.48KK66 pKa = 9.6TSSIKK71 pKa = 8.59VHH73 pKa = 6.31RR74 pKa = 11.84LVAFSFCSGYY84 pKa = 9.2TEE86 pKa = 4.04GLEE89 pKa = 4.25VNHH92 pKa = 7.29KK93 pKa = 10.87DD94 pKa = 4.39GIRR97 pKa = 11.84DD98 pKa = 3.5NNLYY102 pKa = 10.26TNLEE106 pKa = 4.14WVTRR110 pKa = 11.84SQNIRR115 pKa = 11.84DD116 pKa = 3.62TYY118 pKa = 9.56KK119 pKa = 10.43RR120 pKa = 11.84GRR122 pKa = 11.84ITYY125 pKa = 7.32GQKK128 pKa = 10.74NNASKK133 pKa = 11.07LMDD136 pKa = 4.12RR137 pKa = 11.84DD138 pKa = 3.12IGVISSLYY146 pKa = 10.48DD147 pKa = 3.19SGVSQSIISLAFGVSQSTISNVIKK171 pKa = 10.62NKK173 pKa = 9.74HH174 pKa = 5.46YY175 pKa = 11.08KK176 pKa = 10.11NGLIEE181 pKa = 5.52KK182 pKa = 10.11GLADD186 pKa = 3.63QQRR189 pKa = 11.84TNN191 pKa = 3.23
Molecular weight: 21.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
23542 |
33 |
1871 |
182.5 |
20.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.944 ± 0.265 | 1.058 ± 0.137 |
6.448 ± 0.177 | 7.616 ± 0.389 |
4.333 ± 0.194 | 5.926 ± 0.241 |
1.355 ± 0.169 | 7.897 ± 0.249 |
9.162 ± 0.44 | 8.168 ± 0.212 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.268 ± 0.16 | 6.92 ± 0.252 |
2.646 ± 0.131 | 3.25 ± 0.188 |
3.53 ± 0.18 | 7.051 ± 0.235 |
6.525 ± 0.32 | 5.569 ± 0.156 |
1.053 ± 0.077 | 4.282 ± 0.201 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
