
Wenling narna-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.52
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KII8|A0A1L3KII8_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 5 OX=1923505 PE=4 SV=1
MM1 pKa = 7.36EE2 pKa = 5.62AVTSQASALTHH13 pKa = 5.17VQLVSSIKK21 pKa = 10.41YY22 pKa = 9.48YY23 pKa = 9.75STLYY27 pKa = 8.01MCKK30 pKa = 8.24MTNTPFDD37 pKa = 3.79EE38 pKa = 4.6PAQPRR43 pKa = 11.84FHH45 pKa = 6.73SCRR48 pKa = 11.84LHH50 pKa = 6.24RR51 pKa = 11.84HH52 pKa = 5.81LLRR55 pKa = 11.84CSFNPSMKK63 pKa = 9.26QRR65 pKa = 11.84RR66 pKa = 11.84MLVSLLMVKK75 pKa = 10.19KK76 pKa = 10.58GYY78 pKa = 10.62GLAPEE83 pKa = 5.01DD84 pKa = 3.61MVEE87 pKa = 4.05NSLSEE92 pKa = 3.99HH93 pKa = 6.23AAVLGTPGIIADD105 pKa = 4.03GQISDD110 pKa = 5.11GILDD114 pKa = 3.82LVEE117 pKa = 4.46GLFPVGWDD125 pKa = 3.26KK126 pKa = 11.33HH127 pKa = 4.65GHH129 pKa = 5.78CGSLQSRR136 pKa = 11.84AVYY139 pKa = 10.05EE140 pKa = 4.27GGVATNHH147 pKa = 5.74VGSVGLPAFGDD158 pKa = 3.33ACFNYY163 pKa = 10.17NRR165 pKa = 11.84FLCMEE170 pKa = 4.33RR171 pKa = 11.84HH172 pKa = 5.67YY173 pKa = 10.81KK174 pKa = 10.2VKK176 pKa = 10.54ACAVQQPGKK185 pKa = 10.35IRR187 pKa = 11.84IITKK191 pKa = 10.34GEE193 pKa = 3.9VSLKK197 pKa = 10.67CLMPLQNSIMAQLRR211 pKa = 11.84KK212 pKa = 10.07NKK214 pKa = 9.95SFCLTNRR221 pKa = 11.84EE222 pKa = 4.22LEE224 pKa = 4.19KK225 pKa = 10.57TDD227 pKa = 4.03IPKK230 pKa = 10.29ADD232 pKa = 3.87PKK234 pKa = 10.94RR235 pKa = 11.84PFVSGDD241 pKa = 3.3YY242 pKa = 10.69KK243 pKa = 11.15SATDD247 pKa = 3.68YY248 pKa = 11.22LCPEE252 pKa = 4.17LQYY255 pKa = 11.45NIMRR259 pKa = 11.84VILEE263 pKa = 4.11RR264 pKa = 11.84STSEE268 pKa = 3.8WVKK271 pKa = 10.76KK272 pKa = 10.12LWWTALEE279 pKa = 4.0EE280 pKa = 4.59VSCHH284 pKa = 5.65EE285 pKa = 3.81IEE287 pKa = 4.52YY288 pKa = 10.16PNGTSIHH295 pKa = 4.93QARR298 pKa = 11.84GQLMGSLLSFPLLCIINYY316 pKa = 9.47CVLQYY321 pKa = 10.91LFKK324 pKa = 11.17GRR326 pKa = 11.84WCLINGDD333 pKa = 4.92DD334 pKa = 5.82ILFQANKK341 pKa = 9.91QEE343 pKa = 3.89YY344 pKa = 9.48SLWCEE349 pKa = 3.55TVARR353 pKa = 11.84VGFQLSLGKK362 pKa = 10.23NYY364 pKa = 7.99HH365 pKa = 4.88TNKK368 pKa = 10.39LYY370 pKa = 10.43TINSRR375 pKa = 11.84FFKK378 pKa = 11.26YY379 pKa = 10.51NFLKK383 pKa = 9.56NTHH386 pKa = 6.67DD387 pKa = 4.51EE388 pKa = 3.98ICFLHH393 pKa = 6.93LGRR396 pKa = 11.84MFGPMSGDD404 pKa = 3.34TYY406 pKa = 11.18RR407 pKa = 11.84RR408 pKa = 11.84IPSYY412 pKa = 8.85YY413 pKa = 7.57QHH415 pKa = 6.43MYY417 pKa = 10.32KK418 pKa = 10.25RR419 pKa = 11.84YY420 pKa = 8.1CKK422 pKa = 9.9KK423 pKa = 10.09VPHH426 pKa = 6.21MRR428 pKa = 11.84DD429 pKa = 3.04LLEE432 pKa = 4.14PVCYY436 pKa = 10.26GGMGGVTFDD445 pKa = 4.83EE446 pKa = 4.49YY447 pKa = 11.68LKK449 pKa = 9.85MVPVRR454 pKa = 11.84RR455 pKa = 11.84GSSLALHH462 pKa = 6.2HH463 pKa = 5.86QLVRR467 pKa = 11.84LGTPRR472 pKa = 11.84LAVMAYY478 pKa = 10.19DD479 pKa = 3.68VLVSLWFGWLTWSIPTCRR497 pKa = 11.84RR498 pKa = 11.84PEE500 pKa = 3.89HH501 pKa = 6.81PIGGFPRR508 pKa = 11.84VKK510 pKa = 10.63GKK512 pKa = 10.29LSCLLEE518 pKa = 4.06PRR520 pKa = 11.84MKK522 pKa = 10.12PWAPNVLEE530 pKa = 5.0FIRR533 pKa = 5.2
MM1 pKa = 7.36EE2 pKa = 5.62AVTSQASALTHH13 pKa = 5.17VQLVSSIKK21 pKa = 10.41YY22 pKa = 9.48YY23 pKa = 9.75STLYY27 pKa = 8.01MCKK30 pKa = 8.24MTNTPFDD37 pKa = 3.79EE38 pKa = 4.6PAQPRR43 pKa = 11.84FHH45 pKa = 6.73SCRR48 pKa = 11.84LHH50 pKa = 6.24RR51 pKa = 11.84HH52 pKa = 5.81LLRR55 pKa = 11.84CSFNPSMKK63 pKa = 9.26QRR65 pKa = 11.84RR66 pKa = 11.84MLVSLLMVKK75 pKa = 10.19KK76 pKa = 10.58GYY78 pKa = 10.62GLAPEE83 pKa = 5.01DD84 pKa = 3.61MVEE87 pKa = 4.05NSLSEE92 pKa = 3.99HH93 pKa = 6.23AAVLGTPGIIADD105 pKa = 4.03GQISDD110 pKa = 5.11GILDD114 pKa = 3.82LVEE117 pKa = 4.46GLFPVGWDD125 pKa = 3.26KK126 pKa = 11.33HH127 pKa = 4.65GHH129 pKa = 5.78CGSLQSRR136 pKa = 11.84AVYY139 pKa = 10.05EE140 pKa = 4.27GGVATNHH147 pKa = 5.74VGSVGLPAFGDD158 pKa = 3.33ACFNYY163 pKa = 10.17NRR165 pKa = 11.84FLCMEE170 pKa = 4.33RR171 pKa = 11.84HH172 pKa = 5.67YY173 pKa = 10.81KK174 pKa = 10.2VKK176 pKa = 10.54ACAVQQPGKK185 pKa = 10.35IRR187 pKa = 11.84IITKK191 pKa = 10.34GEE193 pKa = 3.9VSLKK197 pKa = 10.67CLMPLQNSIMAQLRR211 pKa = 11.84KK212 pKa = 10.07NKK214 pKa = 9.95SFCLTNRR221 pKa = 11.84EE222 pKa = 4.22LEE224 pKa = 4.19KK225 pKa = 10.57TDD227 pKa = 4.03IPKK230 pKa = 10.29ADD232 pKa = 3.87PKK234 pKa = 10.94RR235 pKa = 11.84PFVSGDD241 pKa = 3.3YY242 pKa = 10.69KK243 pKa = 11.15SATDD247 pKa = 3.68YY248 pKa = 11.22LCPEE252 pKa = 4.17LQYY255 pKa = 11.45NIMRR259 pKa = 11.84VILEE263 pKa = 4.11RR264 pKa = 11.84STSEE268 pKa = 3.8WVKK271 pKa = 10.76KK272 pKa = 10.12LWWTALEE279 pKa = 4.0EE280 pKa = 4.59VSCHH284 pKa = 5.65EE285 pKa = 3.81IEE287 pKa = 4.52YY288 pKa = 10.16PNGTSIHH295 pKa = 4.93QARR298 pKa = 11.84GQLMGSLLSFPLLCIINYY316 pKa = 9.47CVLQYY321 pKa = 10.91LFKK324 pKa = 11.17GRR326 pKa = 11.84WCLINGDD333 pKa = 4.92DD334 pKa = 5.82ILFQANKK341 pKa = 9.91QEE343 pKa = 3.89YY344 pKa = 9.48SLWCEE349 pKa = 3.55TVARR353 pKa = 11.84VGFQLSLGKK362 pKa = 10.23NYY364 pKa = 7.99HH365 pKa = 4.88TNKK368 pKa = 10.39LYY370 pKa = 10.43TINSRR375 pKa = 11.84FFKK378 pKa = 11.26YY379 pKa = 10.51NFLKK383 pKa = 9.56NTHH386 pKa = 6.67DD387 pKa = 4.51EE388 pKa = 3.98ICFLHH393 pKa = 6.93LGRR396 pKa = 11.84MFGPMSGDD404 pKa = 3.34TYY406 pKa = 11.18RR407 pKa = 11.84RR408 pKa = 11.84IPSYY412 pKa = 8.85YY413 pKa = 7.57QHH415 pKa = 6.43MYY417 pKa = 10.32KK418 pKa = 10.25RR419 pKa = 11.84YY420 pKa = 8.1CKK422 pKa = 9.9KK423 pKa = 10.09VPHH426 pKa = 6.21MRR428 pKa = 11.84DD429 pKa = 3.04LLEE432 pKa = 4.14PVCYY436 pKa = 10.26GGMGGVTFDD445 pKa = 4.83EE446 pKa = 4.49YY447 pKa = 11.68LKK449 pKa = 9.85MVPVRR454 pKa = 11.84RR455 pKa = 11.84GSSLALHH462 pKa = 6.2HH463 pKa = 5.86QLVRR467 pKa = 11.84LGTPRR472 pKa = 11.84LAVMAYY478 pKa = 10.19DD479 pKa = 3.68VLVSLWFGWLTWSIPTCRR497 pKa = 11.84RR498 pKa = 11.84PEE500 pKa = 3.89HH501 pKa = 6.81PIGGFPRR508 pKa = 11.84VKK510 pKa = 10.63GKK512 pKa = 10.29LSCLLEE518 pKa = 4.06PRR520 pKa = 11.84MKK522 pKa = 10.12PWAPNVLEE530 pKa = 5.0FIRR533 pKa = 5.2
Molecular weight: 60.99 kDa
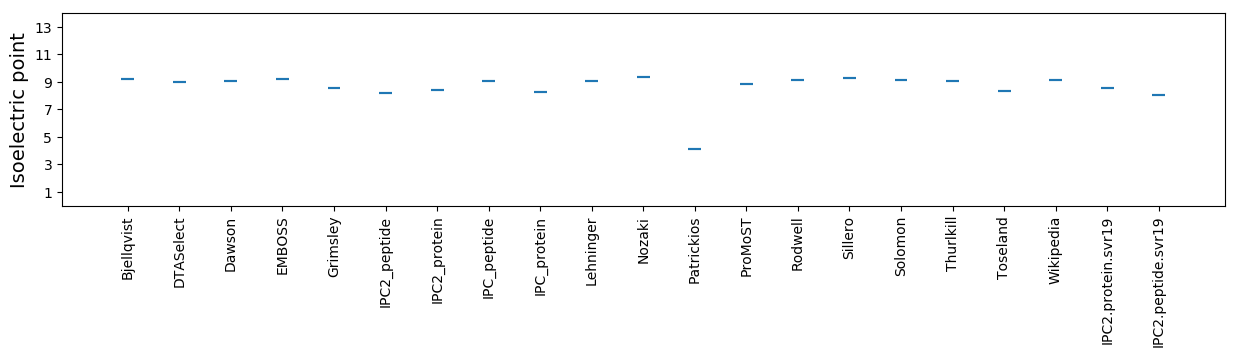
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KII8|A0A1L3KII8_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 5 OX=1923505 PE=4 SV=1
MM1 pKa = 7.36EE2 pKa = 5.62AVTSQASALTHH13 pKa = 5.17VQLVSSIKK21 pKa = 10.41YY22 pKa = 9.48YY23 pKa = 9.75STLYY27 pKa = 8.01MCKK30 pKa = 8.24MTNTPFDD37 pKa = 3.79EE38 pKa = 4.6PAQPRR43 pKa = 11.84FHH45 pKa = 6.73SCRR48 pKa = 11.84LHH50 pKa = 6.24RR51 pKa = 11.84HH52 pKa = 5.81LLRR55 pKa = 11.84CSFNPSMKK63 pKa = 9.26QRR65 pKa = 11.84RR66 pKa = 11.84MLVSLLMVKK75 pKa = 10.19KK76 pKa = 10.58GYY78 pKa = 10.62GLAPEE83 pKa = 5.01DD84 pKa = 3.61MVEE87 pKa = 4.05NSLSEE92 pKa = 3.99HH93 pKa = 6.23AAVLGTPGIIADD105 pKa = 4.03GQISDD110 pKa = 5.11GILDD114 pKa = 3.82LVEE117 pKa = 4.46GLFPVGWDD125 pKa = 3.26KK126 pKa = 11.33HH127 pKa = 4.65GHH129 pKa = 5.78CGSLQSRR136 pKa = 11.84AVYY139 pKa = 10.05EE140 pKa = 4.27GGVATNHH147 pKa = 5.74VGSVGLPAFGDD158 pKa = 3.33ACFNYY163 pKa = 10.17NRR165 pKa = 11.84FLCMEE170 pKa = 4.33RR171 pKa = 11.84HH172 pKa = 5.67YY173 pKa = 10.81KK174 pKa = 10.2VKK176 pKa = 10.54ACAVQQPGKK185 pKa = 10.35IRR187 pKa = 11.84IITKK191 pKa = 10.34GEE193 pKa = 3.9VSLKK197 pKa = 10.67CLMPLQNSIMAQLRR211 pKa = 11.84KK212 pKa = 10.07NKK214 pKa = 9.95SFCLTNRR221 pKa = 11.84EE222 pKa = 4.22LEE224 pKa = 4.19KK225 pKa = 10.57TDD227 pKa = 4.03IPKK230 pKa = 10.29ADD232 pKa = 3.87PKK234 pKa = 10.94RR235 pKa = 11.84PFVSGDD241 pKa = 3.3YY242 pKa = 10.69KK243 pKa = 11.15SATDD247 pKa = 3.68YY248 pKa = 11.22LCPEE252 pKa = 4.17LQYY255 pKa = 11.45NIMRR259 pKa = 11.84VILEE263 pKa = 4.11RR264 pKa = 11.84STSEE268 pKa = 3.8WVKK271 pKa = 10.76KK272 pKa = 10.12LWWTALEE279 pKa = 4.0EE280 pKa = 4.59VSCHH284 pKa = 5.65EE285 pKa = 3.81IEE287 pKa = 4.52YY288 pKa = 10.16PNGTSIHH295 pKa = 4.93QARR298 pKa = 11.84GQLMGSLLSFPLLCIINYY316 pKa = 9.47CVLQYY321 pKa = 10.91LFKK324 pKa = 11.17GRR326 pKa = 11.84WCLINGDD333 pKa = 4.92DD334 pKa = 5.82ILFQANKK341 pKa = 9.91QEE343 pKa = 3.89YY344 pKa = 9.48SLWCEE349 pKa = 3.55TVARR353 pKa = 11.84VGFQLSLGKK362 pKa = 10.23NYY364 pKa = 7.99HH365 pKa = 4.88TNKK368 pKa = 10.39LYY370 pKa = 10.43TINSRR375 pKa = 11.84FFKK378 pKa = 11.26YY379 pKa = 10.51NFLKK383 pKa = 9.56NTHH386 pKa = 6.67DD387 pKa = 4.51EE388 pKa = 3.98ICFLHH393 pKa = 6.93LGRR396 pKa = 11.84MFGPMSGDD404 pKa = 3.34TYY406 pKa = 11.18RR407 pKa = 11.84RR408 pKa = 11.84IPSYY412 pKa = 8.85YY413 pKa = 7.57QHH415 pKa = 6.43MYY417 pKa = 10.32KK418 pKa = 10.25RR419 pKa = 11.84YY420 pKa = 8.1CKK422 pKa = 9.9KK423 pKa = 10.09VPHH426 pKa = 6.21MRR428 pKa = 11.84DD429 pKa = 3.04LLEE432 pKa = 4.14PVCYY436 pKa = 10.26GGMGGVTFDD445 pKa = 4.83EE446 pKa = 4.49YY447 pKa = 11.68LKK449 pKa = 9.85MVPVRR454 pKa = 11.84RR455 pKa = 11.84GSSLALHH462 pKa = 6.2HH463 pKa = 5.86QLVRR467 pKa = 11.84LGTPRR472 pKa = 11.84LAVMAYY478 pKa = 10.19DD479 pKa = 3.68VLVSLWFGWLTWSIPTCRR497 pKa = 11.84RR498 pKa = 11.84PEE500 pKa = 3.89HH501 pKa = 6.81PIGGFPRR508 pKa = 11.84VKK510 pKa = 10.63GKK512 pKa = 10.29LSCLLEE518 pKa = 4.06PRR520 pKa = 11.84MKK522 pKa = 10.12PWAPNVLEE530 pKa = 5.0FIRR533 pKa = 5.2
MM1 pKa = 7.36EE2 pKa = 5.62AVTSQASALTHH13 pKa = 5.17VQLVSSIKK21 pKa = 10.41YY22 pKa = 9.48YY23 pKa = 9.75STLYY27 pKa = 8.01MCKK30 pKa = 8.24MTNTPFDD37 pKa = 3.79EE38 pKa = 4.6PAQPRR43 pKa = 11.84FHH45 pKa = 6.73SCRR48 pKa = 11.84LHH50 pKa = 6.24RR51 pKa = 11.84HH52 pKa = 5.81LLRR55 pKa = 11.84CSFNPSMKK63 pKa = 9.26QRR65 pKa = 11.84RR66 pKa = 11.84MLVSLLMVKK75 pKa = 10.19KK76 pKa = 10.58GYY78 pKa = 10.62GLAPEE83 pKa = 5.01DD84 pKa = 3.61MVEE87 pKa = 4.05NSLSEE92 pKa = 3.99HH93 pKa = 6.23AAVLGTPGIIADD105 pKa = 4.03GQISDD110 pKa = 5.11GILDD114 pKa = 3.82LVEE117 pKa = 4.46GLFPVGWDD125 pKa = 3.26KK126 pKa = 11.33HH127 pKa = 4.65GHH129 pKa = 5.78CGSLQSRR136 pKa = 11.84AVYY139 pKa = 10.05EE140 pKa = 4.27GGVATNHH147 pKa = 5.74VGSVGLPAFGDD158 pKa = 3.33ACFNYY163 pKa = 10.17NRR165 pKa = 11.84FLCMEE170 pKa = 4.33RR171 pKa = 11.84HH172 pKa = 5.67YY173 pKa = 10.81KK174 pKa = 10.2VKK176 pKa = 10.54ACAVQQPGKK185 pKa = 10.35IRR187 pKa = 11.84IITKK191 pKa = 10.34GEE193 pKa = 3.9VSLKK197 pKa = 10.67CLMPLQNSIMAQLRR211 pKa = 11.84KK212 pKa = 10.07NKK214 pKa = 9.95SFCLTNRR221 pKa = 11.84EE222 pKa = 4.22LEE224 pKa = 4.19KK225 pKa = 10.57TDD227 pKa = 4.03IPKK230 pKa = 10.29ADD232 pKa = 3.87PKK234 pKa = 10.94RR235 pKa = 11.84PFVSGDD241 pKa = 3.3YY242 pKa = 10.69KK243 pKa = 11.15SATDD247 pKa = 3.68YY248 pKa = 11.22LCPEE252 pKa = 4.17LQYY255 pKa = 11.45NIMRR259 pKa = 11.84VILEE263 pKa = 4.11RR264 pKa = 11.84STSEE268 pKa = 3.8WVKK271 pKa = 10.76KK272 pKa = 10.12LWWTALEE279 pKa = 4.0EE280 pKa = 4.59VSCHH284 pKa = 5.65EE285 pKa = 3.81IEE287 pKa = 4.52YY288 pKa = 10.16PNGTSIHH295 pKa = 4.93QARR298 pKa = 11.84GQLMGSLLSFPLLCIINYY316 pKa = 9.47CVLQYY321 pKa = 10.91LFKK324 pKa = 11.17GRR326 pKa = 11.84WCLINGDD333 pKa = 4.92DD334 pKa = 5.82ILFQANKK341 pKa = 9.91QEE343 pKa = 3.89YY344 pKa = 9.48SLWCEE349 pKa = 3.55TVARR353 pKa = 11.84VGFQLSLGKK362 pKa = 10.23NYY364 pKa = 7.99HH365 pKa = 4.88TNKK368 pKa = 10.39LYY370 pKa = 10.43TINSRR375 pKa = 11.84FFKK378 pKa = 11.26YY379 pKa = 10.51NFLKK383 pKa = 9.56NTHH386 pKa = 6.67DD387 pKa = 4.51EE388 pKa = 3.98ICFLHH393 pKa = 6.93LGRR396 pKa = 11.84MFGPMSGDD404 pKa = 3.34TYY406 pKa = 11.18RR407 pKa = 11.84RR408 pKa = 11.84IPSYY412 pKa = 8.85YY413 pKa = 7.57QHH415 pKa = 6.43MYY417 pKa = 10.32KK418 pKa = 10.25RR419 pKa = 11.84YY420 pKa = 8.1CKK422 pKa = 9.9KK423 pKa = 10.09VPHH426 pKa = 6.21MRR428 pKa = 11.84DD429 pKa = 3.04LLEE432 pKa = 4.14PVCYY436 pKa = 10.26GGMGGVTFDD445 pKa = 4.83EE446 pKa = 4.49YY447 pKa = 11.68LKK449 pKa = 9.85MVPVRR454 pKa = 11.84RR455 pKa = 11.84GSSLALHH462 pKa = 6.2HH463 pKa = 5.86QLVRR467 pKa = 11.84LGTPRR472 pKa = 11.84LAVMAYY478 pKa = 10.19DD479 pKa = 3.68VLVSLWFGWLTWSIPTCRR497 pKa = 11.84RR498 pKa = 11.84PEE500 pKa = 3.89HH501 pKa = 6.81PIGGFPRR508 pKa = 11.84VKK510 pKa = 10.63GKK512 pKa = 10.29LSCLLEE518 pKa = 4.06PRR520 pKa = 11.84MKK522 pKa = 10.12PWAPNVLEE530 pKa = 5.0FIRR533 pKa = 5.2
Molecular weight: 60.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
533 |
533 |
533 |
533.0 |
60.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.69 ± 0.0 | 3.752 ± 0.0 |
3.377 ± 0.0 | 4.878 ± 0.0 |
4.128 ± 0.0 | 7.129 ± 0.0 |
3.565 ± 0.0 | 4.503 ± 0.0 |
6.004 ± 0.0 | 11.257 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.752 ± 0.0 | 3.752 ± 0.0 |
5.441 ± 0.0 | 3.565 ± 0.0 |
6.191 ± 0.0 | 6.754 ± 0.0 |
4.315 ± 0.0 | 6.379 ± 0.0 |
1.876 ± 0.0 | 4.69 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
